Pre Gene Modal
BGIBMGA007065
Annotation
PREDICTED:_nucleolin_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.196
Sequence
CDS
ATGGACACAGTGCATGTGCTTGTGAAATACGAGATGAGGTCGTTGTTCTATTTGGCGGCGTTATGCTGTCTCGTTTTGTCAGCCTACGGGAATCCTATCTCGCGGCCTGAAGAGGATAAGGAAGTGGGGAGGCAGGCCGTCGCTCCAGCCGCTGTAGCGCCAGCCGCGTCCGCTGATGACGACGACGACGACGACGATGATGACGATGACGATGATGATGATGTTGGTCTTGATTTGCCAGTAGATGACGACGATGAAGACGATGATGAAGACGACGAAGAAGAAGCTGATGATGATGAAGACGATGACTACCTGGAAAGGATTTTCGATGAAATTTTAGGGGGAGACGAAGATGATGATGACGAACCAGCTGTAGCAGCTGTACAGCCTGTAGCAGCCCCAGTGGATACTGCAGCAGCAGCACCCGTAGCTGCTGTTGCTGAAGAAGCTGCCGCTGCTGCTGAAGACGTCGAAGCATCAGAAACCGAAACCGTCGCTGAAGGCGAAGAAAACGAGGTGCAAAAGGATCCCATTAGTGCTGAGACAGAAGCCGTTGCAGCTAGTTCAGAAGCTGCCGCTGCCACCGAAGCCGAAGACGACGACGACGACGAAGATGACATCGACGACGCTCTCGAACCAGAAGATGATGATGAGGAAGACGAGGAAGAAGATGACGACGAAGAAGACGATGTCACTGACGCTTTCGGCAGAAATGTCTCGGCTAAGCACTCACGCGCCTTGAGTGATGAACCGAACAAGAAGCCTGCTCCTGAGGCTGCGGCGCACAACGATATTTTCAAAAGATATAACAGATTCGTAGATAATATCCTGCAGAAGATGAACGACATTTTAAGGAAGTCCTACGACCCAGTGAACGTAAAGCTACAGCCGATAGACGCAAGTAAAAAAAGTAATAAACCCAAAAAGAACAAACCAAAATCAAATAAAAGGAAAACAAACAAAAAGAACACATCTCGACGAAATTCGACTAAGAAAAATAAAGTTAAGTCAACTGAAATCCCTGAAACTAGAAAAGAAGAAATTGTAGAAAATAGCGATGTTAAGCCTATCGAAAATGTCATTGAGGAAATTACCAATGCTACATCTGAATCTAGAGCTATTAAAGACACAAAGGCTAAGCCGACAACTATTAAAGCTAAACCGCCAACGAAAGCCACTAAACCAAAACCCAAGCCCCCAACAAAGACAACAACAAAGAAACCAGTCACAACAAAGAACAAGACAAAGACAAGTGAAAAAAGCAGACCGAGAGCCAAAGGGACTCTATACGGATTATCTTCTTTGAGGAGAAAGGGAGACGTTTCTGTCAAAATAGAGGCAGATCACACGACTGTTAAAAGTAATTTCGCAGTTGGACCTCTTGTTTTAAGAGTCGAGAAAGAGGTCGGACGTGGAGCTAAGAAAGAGTTGAAATCTGCCACCGCCACCACGGAGGAGATGATCGGGAAATTGAGTTTCAGAGTTAGTAGCCAGGGTGTCGCCACACTGCATTCGATCAAGGTTCTACAGCCGAAACAGGTACGAGTCGTCAGTAATCACGAGCGCACTCGTGAGTTAGTATGGCAACGAAGCTCAAGAATAGCTCACGTCGTCTCTGAAAAGTTGTTGTCCGCATCTAGACCTATGTTCAACCAACATACTGTAGTAAAACAGTAA
Protein
MDTVHVLVKYEMRSLFYLAALCCLVLSAYGNPISRPEEDKEVGRQAVAPAAVAPAASADDDDDDDDDDDDDDDDVGLDLPVDDDDEDDDEDDEEEADDDEDDDYLERIFDEILGGDEDDDDEPAVAAVQPVAAPVDTAAAAPVAAVAEEAAAAAEDVEASETETVAEGEENEVQKDPISAETEAVAASSEAAAATEAEDDDDDEDDIDDALEPEDDDEEDEEEDDDEEDDVTDAFGRNVSAKHSRALSDEPNKKPAPEAAAHNDIFKRYNRFVDNILQKMNDILRKSYDPVNVKLQPIDASKKSNKPKKNKPKSNKRKTNKKNTSRRNSTKKNKVKSTEIPETRKEEIVENSDVKPIENVIEEITNATSESRAIKDTKAKPTTIKAKPPTKATKPKPKPPTKTTTKKPVTTKNKTKTSEKSRPRAKGTLYGLSSLRRKGDVSVKIEADHTTVKSNFAVGPLVLRVEKEVGRGAKKELKSATATTEEMIGKLSFRVSSQGVATLHSIKVLQPKQVRVVSNHERTRELVWQRSSRIAHVVSEKLLSASRPMFNQHTVVKQ
Summary
Uniprot
ProteinModelPortal
Ontologies
GO
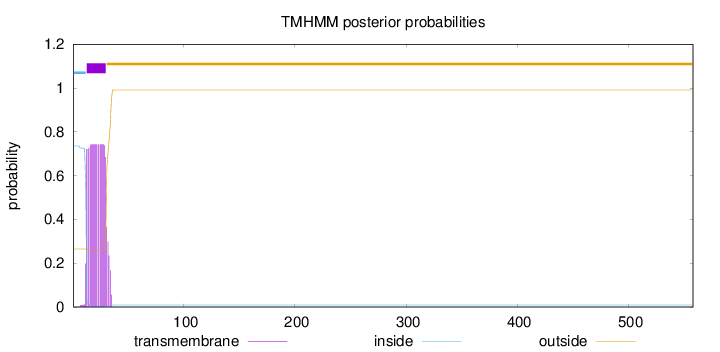
Topology
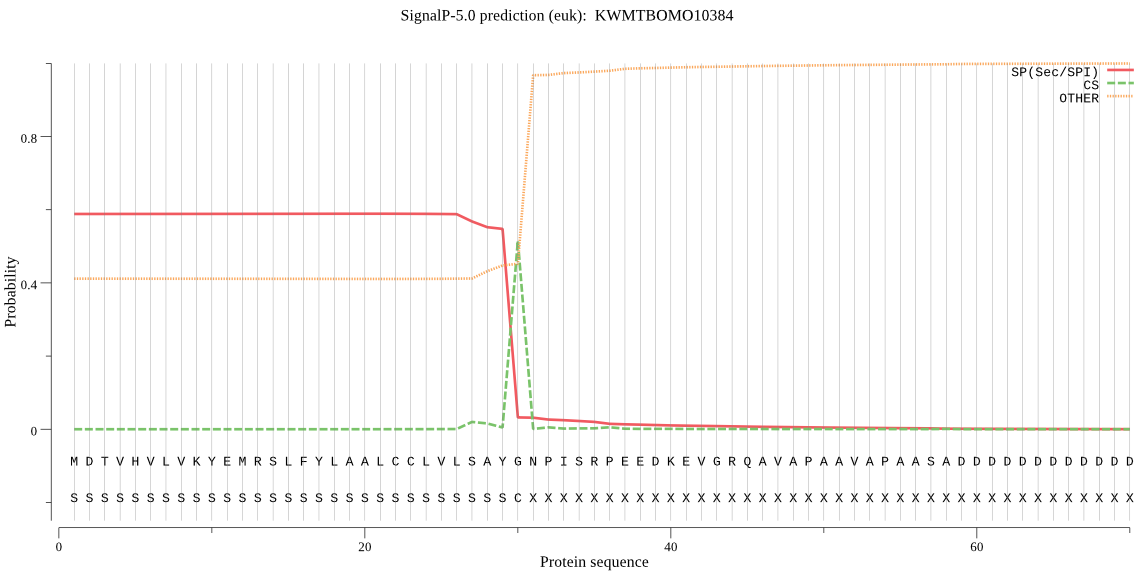
SignalP
Position: 1 - 30,
Likelihood: 0.589651
Length:
558
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
14.62133
Exp number, first 60 AAs:
14.6211
Total prob of N-in:
0.73492
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 30
outside
31 - 558
Population Genetic Test Statistics
Pi
29.106135
Theta
225.582691
Tajima's D
0.80985
CLR
0.001269
CSRT
0.604269786510674
Interpretation
Uncertain
