Gene
KWMTBOMO10383 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007042
Annotation
PREDICTED:_uncharacterized_protein_LOC101742755_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.556 PlasmaMembrane Reliability : 1.094
Sequence
CDS
ATGGTCAACGAAGCTCTCGCTTTGGAACGAAGATTCTTATTAGGCGCTAATGATGCATCAAACTGTACGTACGATGATGAGATCAGCAACATCCCTTGTCCACCCAGCAAGTATCGCAGCGCGAGCGGAGAATGCAACAACGTTCGACATAGGCCCTGGGGTAGAAGAGGAGATATATTCTTACGGTTGATGCCACCTAATTACGCTAATGATGTATCGCTGCCTCTGTCAACGCCTCATCTGCCGGAGCCGTCGCTGGCCATCCGTGCGGCGGCACACTTGTCTCTGCCAGCCGACCATGACTTCGTGACCAGTATGCTGGCTGCCTGGGGGCAATTTCTTATCGACGATCTCGTTGAAACTTCTAATGGCGATAAGGACTGCGGCGATTCCTATTGCGAGTACATACGCTCTGCGCCAACGAGAGACATCGGTTCATGTGGCTTTGAGCACCGCGAACAGATGAACTTGGCTACATCTTTCCTCGACGGTTCCGCGTTATACGGAAGCTCGGAGAAGGAACTGCGGTCGCTACGTCTATACGACGCTGGGAAAGTTGATATAGCCTCGTGCAGAAAATGCAACATTCAGACACCGACAGCTCCTCTGTACAAAGCTCTTTTATCGGAGCACAACCGCATCGCCGGACAACTATACTCTTTGAATCCCTTCTGGGACGACACCACCTTGTTCTTGGAAACGAGACGTGCTATGGCCGCTATCATTCAGCATATCACCTACAATGAGTTCTTGCCGGTATTACTAGGCGAGGTGGGAATGGCCAAAGCAGAATTGAAATTGACTTCCCTCGGATTTTGGCGAGGCTACTCAAGTGCCAACCGCGCAGGAATCTACACGGAACTGGTGGCGGTAGCGCCTGTTTTCAAGGCGATGATGAATCAAAAATTGGTCAACACTACGATCTCTCTTGAACATTTAATCCACACCAGCGCCCAACACGTCTCAAGATTCCCTCCCTCAACACAATGGGACCTGAACAGATCAAGAGATCACGGTGTTGCCCCATATTTGAGGGCCTTAAAAATGTGCGAACCGACAGCTGCTGTGAAGACATTCGGAGACTTGGAGAAAATGGGATTTGATCAGACCCAGCAAGAGATGATGGCTGATTTGTACAGGAACGCTGAAGACATGGAATTAATGATAGGCGGTGCAATGGAGAAACCGGCGTCCGGTGCTGTCGTGGGCTCAACACTCGCTTGTGTATTAGCGTTGCAGTTTGCCAACATGAAGAAAAGCGACAGATTTTGGTATGAAAATGATATTCCTCCTTCATCGTTCGCACCGGAACAACTGGCCGCCATCAGGAAAGTTTCTTTGTCCGGCTTACTCTGTGCAGCTGACGACACGATTGTGAACATACAACCTAAGGCTTTTGTTAAGGAAGATCCCTTCTTAAACGCTGCCCAGCATTGCTCCCAACACAATCGTTTGGAACTCGCCGCCTGGAGAGACGAAACTGGCGCCAAAGCAGCTGAACGTTTATCCCACGATATGTTGAACGCCGCTATTGAAAAAGCTAAGCAGGAAATGATCGACAGAAAGAAACTGGAATATATGCTGTGGGAATCACGTGGAGGTGCAGACCCTAAGTCCCCGGTCGGTACAGCCGCGTCGTTCTCAAAAGCCAACAAATATGCTTTGAAACTGGCCAACACTTCGCTGTTCTTGGAGTTTGCTACTAATGAACTGATCAACTCCGTTGGGGAAGGGCATCGTCGCAAACGTCAAATCTTCGACGACGCTCTCGGGTTCGGCACGACCAACGATTTCGCTGATTCATTGCAGTCGGTCGACATTAGCAGCTTCCTCGGACAAGATCAACTCGGTGGGCCGACAATAGAACCGCAATGTGACGACAGCGGCCCGTGCGACGCTAACAGTCCGTTCAGAACGTACACCGGCTACTGCAACAACTTGAGGAACCCAAACCTCGGGAAGAGTTTAACGACCTTCGCTAGACTGCTGCCACCGGTGTATGAAGACGGAGTCAGTCGTCCCCGCATTAACTCAGTTACCGGAACTCCTTTACCGTCGCCTCGTGTGATATCAACTGTAATCCATCCTGATATATCTAACTTGCATACCCGATATACGCTAATGGTGATGCAGTTCGCACAATTTTTGGACCACGAGCTCACCATGACACCGATTCACAAAGGTTTCCACGAGTCCATACCGGACTGCAGATCTTGCGATTCGCCTCGCACCGTCCATCCTGAATGCAATCCATTCCCAGTACCACGAGGCGACCATTACTATCCAGAAGTCAACGTCACTTCCGGAGAACGACTATGCTTCCCCTTCATGAGAAGTTTGCCAGGACAACAGCAGTTAGGACCACGTGAACAAGTTAATCAGAATACGCCGTACATTGACGGTTCCGTGATTTATGGAGAGAATCCTTGCATATTCCGCAAATTGCGTGGTTTCAATGGACGTTTGAACGCTACAAACAATCCTTTCAACGGTAAAGATTTGTTGCCGCGATCTGACAGCCATCCAGAATGCAAAGCCGCCAGTGGATATTGCTTCATTGCTGGTGACGGCAGAGCTTCAGAGCAACCGGCACTGACAGCGATTCACACCATCTTCCTTAGGGAACATAACCGAATTGTCGAAGGCCTGAGAGGCGTGAACCCACACTGGGATACAGAATTGCTCTTCGAACACGCTCGTCGTATTATGGCTGCCCAGTTCACGCATATAATCTATAATGAATTCCTACCCAGATTGCTATCGTGGAACGCTGTCAACCTTTATGGCCTGAAACTGTTGCCTTCCGGATACTACAAAGAGTACTCTCCTACCTGTAACCCTGCAATCGTGACCGAGTTCGCTACAGCAGCCTTCAGGATTGGACATTCATTACTGCGACCACATCTTCCGCGCCTCTCACCCAACTACCAGCCTGTGGAACCACCAATTCTCCTAAGGGACGGCTTTTTCAGAACCGATATGTTCATGTCTCATCCTCCACTTGTCGACGAGCTTATGCGGGGTCTTTCCTCAACTCCGATGGAAACCCTTGACCAGTTCATCACCGGCGAAGTCACTAACCATCTATTCGAAGACCGTCGTATCCCGTTCTCGGGGATCGATTTAATCGCTCTCAACATCCAAAGAGCTAGGGACCACGGTGTTCCTGGATACAATAACTACAGGGCCCTGTGTAATTTAAAAAGAGCTACCACATTCGATGACTTGGCTAGAGAAATTCCTGATGAGGTTATCGCAAGGTTTAAGAGAATTTATGCAACCGTGGATGATATTGATCTGTTCCCCGGTGGAATGAGTGAAAGGCCACTTCAAGGAGGGCTTATCGGACCGACCTTTGCTTGCATCATTGCTATCCAATTCAGGCAGCTAAGAAAATGCGATCGTTTTTGGTACGAAAACGACAATACGGCTGCTCGTTTTACCGAACAACAACTTGCCGAAATCCGTAAAGCGACTCTTTCGAAGGTGCTCTGTGATAACTTTGACATACCCGGCGACATTCAAAGAGCCGCCTTTGACCTACCCAGCAACTTCCTTAATCCACGTGTACCATGCGCATCTCTACCTAAGTTGGATCTGTCAGCGTGGCGTGAAAGCTCAACGCAGGGCTGTCTCATCGCAGGCCGCTCAGTCGCTGTGGGCGAGTCCGCTTTCCCATCACCGTGTACTTCGTGTATCTGCACTGTAGAAGGAGCACAATGCGCCTCTCTCCGTATCACCGACTGCGCGCAGTTGGTCCGCGAGTGGCCGCGCGAGGCGATCCTGCGCGATGACGTGTGCACCGCGCAGTGTCCCACCCCCGCGCCCGTCACACACCCTCGCCCGCCTCGAAGACAGACCATTAACTTCAAATTCCCCGACCTGACACCCTACATAGCGAAGTGA
Protein
MVNEALALERRFLLGANDASNCTYDDEISNIPCPPSKYRSASGECNNVRHRPWGRRGDIFLRLMPPNYANDVSLPLSTPHLPEPSLAIRAAAHLSLPADHDFVTSMLAAWGQFLIDDLVETSNGDKDCGDSYCEYIRSAPTRDIGSCGFEHREQMNLATSFLDGSALYGSSEKELRSLRLYDAGKVDIASCRKCNIQTPTAPLYKALLSEHNRIAGQLYSLNPFWDDTTLFLETRRAMAAIIQHITYNEFLPVLLGEVGMAKAELKLTSLGFWRGYSSANRAGIYTELVAVAPVFKAMMNQKLVNTTISLEHLIHTSAQHVSRFPPSTQWDLNRSRDHGVAPYLRALKMCEPTAAVKTFGDLEKMGFDQTQQEMMADLYRNAEDMELMIGGAMEKPASGAVVGSTLACVLALQFANMKKSDRFWYENDIPPSSFAPEQLAAIRKVSLSGLLCAADDTIVNIQPKAFVKEDPFLNAAQHCSQHNRLELAAWRDETGAKAAERLSHDMLNAAIEKAKQEMIDRKKLEYMLWESRGGADPKSPVGTAASFSKANKYALKLANTSLFLEFATNELINSVGEGHRRKRQIFDDALGFGTTNDFADSLQSVDISSFLGQDQLGGPTIEPQCDDSGPCDANSPFRTYTGYCNNLRNPNLGKSLTTFARLLPPVYEDGVSRPRINSVTGTPLPSPRVISTVIHPDISNLHTRYTLMVMQFAQFLDHELTMTPIHKGFHESIPDCRSCDSPRTVHPECNPFPVPRGDHYYPEVNVTSGERLCFPFMRSLPGQQQLGPREQVNQNTPYIDGSVIYGENPCIFRKLRGFNGRLNATNNPFNGKDLLPRSDSHPECKAASGYCFIAGDGRASEQPALTAIHTIFLREHNRIVEGLRGVNPHWDTELLFEHARRIMAAQFTHIIYNEFLPRLLSWNAVNLYGLKLLPSGYYKEYSPTCNPAIVTEFATAAFRIGHSLLRPHLPRLSPNYQPVEPPILLRDGFFRTDMFMSHPPLVDELMRGLSSTPMETLDQFITGEVTNHLFEDRRIPFSGIDLIALNIQRARDHGVPGYNNYRALCNLKRATTFDDLAREIPDEVIARFKRIYATVDDIDLFPGGMSERPLQGGLIGPTFACIIAIQFRQLRKCDRFWYENDNTAARFTEQQLAEIRKATLSKVLCDNFDIPGDIQRAAFDLPSNFLNPRVPCASLPKLDLSAWRESSTQGCLIAGRSVAVGESAFPSPCTSCICTVEGAQCASLRITDCAQLVREWPREAILRDDVCTAQCPTPAPVTHPRPPRRQTINFKFPDLTPYIAK
Summary
Cofactor
Mg(2+)
Similarity
Belongs to the PP2C family.
Uniprot
H9JBZ7
A0A2H1V903
A0A0N1IPH7
A0A3S2LDE4
A0A194PPD9
A0A212FP75
+ More
A0A2A4J6A6 A0A1Y1K115 A0A182RDN6 A0A182PUE0 A0A182Y399 A0A182QXR0 A0A182JK69 A0A182L0N8 A0A182NTX0 Q7Q4D6 A0A1S4GY16 A0A182UMG8 A0A2M3YZ06 A0A2M3YZ58 A0A2M4B9C9 A0A084W5U2 A0A2M4CTY0 A0A182LTI9 A0A1J1I1B5 A0A2M4CTF6 A0A2M4CTI2 A0A182K1Q9 A0A182W6I3 B0WXB4 D6WXZ9 A0A023EWS4 A0A182H8D0 Q0IFX2 A0A182IC95 A0A182XLC5 A0A1W4XF82 B4MZG3 A0A1I8N7R6 T1PE43 B4KK29 A0A1A9VXM6 A0A034VC57 A0A0M4E6Z3 A0A0K8VET0 A0A0A1XK54 B4LTJ0 Q29MP6 A0A0A1WM92 A0A1I8P8B2 A0A3B0K071 A0A0L0CBR6 B3MM24 A0A310SLN5 B4JDM1 A0A1B0DMT2 A0A158NFQ1 E2C4H1 A0A195BDL5 F4WGS9 A0A1W4V9M9 B4P9U9 Q9VJ80 A0A151JVI7 B4Q8U8 A0A154PPW0 A0A1B0FN29 B4I5H5 A0A1A9ZIV4 A0A0J9R404 A0A151WP86 B3NMA9 A0A195DP72 A0A3L8DBM2 A0A1B0B4N1 A0A1A9XDR7 A0A087ZYF6 A0A0Q9WNC9 U4U893 A0A2J7RIR8 A0A0P9BMS4 A0A0N0BCF0 A0A1A9W0L4 A0A023EWT3 A0A151I936 E9IGL4 N6TXU1 E2AT66 A0A2M4ALX6 A0A0L7R2Q0 A0A023EWS2 A0A2P8Y122 A0A0T6B7C2 E0VHH2 A0A336LIY5 A0A336LN33 A0A023EX84 A0A146MDQ2 A0A0A9VVR2
A0A2A4J6A6 A0A1Y1K115 A0A182RDN6 A0A182PUE0 A0A182Y399 A0A182QXR0 A0A182JK69 A0A182L0N8 A0A182NTX0 Q7Q4D6 A0A1S4GY16 A0A182UMG8 A0A2M3YZ06 A0A2M3YZ58 A0A2M4B9C9 A0A084W5U2 A0A2M4CTY0 A0A182LTI9 A0A1J1I1B5 A0A2M4CTF6 A0A2M4CTI2 A0A182K1Q9 A0A182W6I3 B0WXB4 D6WXZ9 A0A023EWS4 A0A182H8D0 Q0IFX2 A0A182IC95 A0A182XLC5 A0A1W4XF82 B4MZG3 A0A1I8N7R6 T1PE43 B4KK29 A0A1A9VXM6 A0A034VC57 A0A0M4E6Z3 A0A0K8VET0 A0A0A1XK54 B4LTJ0 Q29MP6 A0A0A1WM92 A0A1I8P8B2 A0A3B0K071 A0A0L0CBR6 B3MM24 A0A310SLN5 B4JDM1 A0A1B0DMT2 A0A158NFQ1 E2C4H1 A0A195BDL5 F4WGS9 A0A1W4V9M9 B4P9U9 Q9VJ80 A0A151JVI7 B4Q8U8 A0A154PPW0 A0A1B0FN29 B4I5H5 A0A1A9ZIV4 A0A0J9R404 A0A151WP86 B3NMA9 A0A195DP72 A0A3L8DBM2 A0A1B0B4N1 A0A1A9XDR7 A0A087ZYF6 A0A0Q9WNC9 U4U893 A0A2J7RIR8 A0A0P9BMS4 A0A0N0BCF0 A0A1A9W0L4 A0A023EWT3 A0A151I936 E9IGL4 N6TXU1 E2AT66 A0A2M4ALX6 A0A0L7R2Q0 A0A023EWS2 A0A2P8Y122 A0A0T6B7C2 E0VHH2 A0A336LIY5 A0A336LN33 A0A023EX84 A0A146MDQ2 A0A0A9VVR2
Pubmed
19121390
26354079
22118469
28004739
25244985
20966253
+ More
12364791 24438588 18362917 19820115 24945155 26483478 17510324 17994087 25315136 18057021 25348373 25830018 15632085 26108605 21347285 20798317 21719571 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 30249741 23537049 21282665 29403074 20566863 26823975 25401762
12364791 24438588 18362917 19820115 24945155 26483478 17510324 17994087 25315136 18057021 25348373 25830018 15632085 26108605 21347285 20798317 21719571 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 30249741 23537049 21282665 29403074 20566863 26823975 25401762
EMBL
BABH01014717
ODYU01001281
SOQ37281.1
KQ460328
KPJ15657.1
RSAL01000036
+ More
RVE51299.1 KQ459603 KPI93000.1 AGBW02003464 OWR55548.1 NWSH01002964 PCG67208.1 GEZM01100127 JAV53006.1 MF964191 AXE28372.1 AXCN02000807 AAAB01008964 EAA12252.5 GGFM01000759 MBW21510.1 GGFM01000747 MBW21498.1 GGFJ01000496 MBW49637.1 ATLV01020694 KE525305 KFB45586.1 GGFL01004453 MBW68631.1 AXCM01002012 CVRI01000038 CRK94120.1 GGFL01004454 MBW68632.1 GGFL01004452 MBW68630.1 DS232161 EDS36375.1 KQ971362 EFA09156.2 GAPW01000020 JAC13578.1 JXUM01118153 KQ566155 KXJ70361.1 CH477286 EAT44728.2 APCN01000786 CH963920 EDW77748.2 KA647037 AFP61666.1 CH933807 EDW11547.1 KRG02753.1 GAKP01019592 GAKP01019591 JAC39360.1 CP012523 ALC40196.1 GDHF01014855 JAI37459.1 GBXI01002990 JAD11302.1 CH940649 EDW64963.1 CH379060 EAL33647.2 GBXI01014764 JAC99527.1 OUUW01000004 SPP79006.1 JRES01000613 KNC29858.1 CH902620 EDV30839.1 KQ760116 OAD61931.1 CH916368 EDW03391.1 AJVK01007104 AJVK01007105 AJVK01007106 AJVK01007107 ADTU01014496 ADTU01014497 ADTU01014498 ADTU01014499 ADTU01014500 GL452471 EFN77155.1 KQ976514 KYM82275.1 GL888147 EGI66548.1 CM000158 EDW90290.1 AE014134 AY051952 AAF53674.3 AAK93376.1 KQ981724 KYN37231.1 CM000361 EDX05380.1 KQ435022 KZC13921.1 CCAG010022043 CH480822 EDW55631.1 CM002910 KMY90823.1 KQ982877 KYQ49709.1 CH954179 EDV54780.1 KQ980662 KYN14715.1 QOIP01000010 RLU17910.1 JXJN01008376 KRF81957.1 KB632075 ERL88558.1 NEVH01003493 PNF40733.1 KPU73076.1 KPU73077.1 KPU73078.1 KQ435912 KOX68884.1 GAPW01000041 JAC13557.1 KQ978311 KYM95318.1 GL763048 EFZ20307.1 APGK01050727 APGK01050728 APGK01050729 APGK01050730 APGK01050731 APGK01050732 APGK01050733 APGK01050734 APGK01050735 KB741176 ENN73201.1 GL442519 EFN63376.1 GGFK01008426 MBW41747.1 KQ414665 KOC65152.1 GAPW01000061 JAC13537.1 PYGN01001063 PSN37939.1 LJIG01009350 KRT83256.1 DS235170 EEB12828.1 UFQT01000026 SSX18052.1 SSX18053.1 GAPW01000082 JAC13516.1 GDHC01000678 JAQ17951.1 GBHO01044318 GBHO01037253 JAF99285.1 JAG06351.1
RVE51299.1 KQ459603 KPI93000.1 AGBW02003464 OWR55548.1 NWSH01002964 PCG67208.1 GEZM01100127 JAV53006.1 MF964191 AXE28372.1 AXCN02000807 AAAB01008964 EAA12252.5 GGFM01000759 MBW21510.1 GGFM01000747 MBW21498.1 GGFJ01000496 MBW49637.1 ATLV01020694 KE525305 KFB45586.1 GGFL01004453 MBW68631.1 AXCM01002012 CVRI01000038 CRK94120.1 GGFL01004454 MBW68632.1 GGFL01004452 MBW68630.1 DS232161 EDS36375.1 KQ971362 EFA09156.2 GAPW01000020 JAC13578.1 JXUM01118153 KQ566155 KXJ70361.1 CH477286 EAT44728.2 APCN01000786 CH963920 EDW77748.2 KA647037 AFP61666.1 CH933807 EDW11547.1 KRG02753.1 GAKP01019592 GAKP01019591 JAC39360.1 CP012523 ALC40196.1 GDHF01014855 JAI37459.1 GBXI01002990 JAD11302.1 CH940649 EDW64963.1 CH379060 EAL33647.2 GBXI01014764 JAC99527.1 OUUW01000004 SPP79006.1 JRES01000613 KNC29858.1 CH902620 EDV30839.1 KQ760116 OAD61931.1 CH916368 EDW03391.1 AJVK01007104 AJVK01007105 AJVK01007106 AJVK01007107 ADTU01014496 ADTU01014497 ADTU01014498 ADTU01014499 ADTU01014500 GL452471 EFN77155.1 KQ976514 KYM82275.1 GL888147 EGI66548.1 CM000158 EDW90290.1 AE014134 AY051952 AAF53674.3 AAK93376.1 KQ981724 KYN37231.1 CM000361 EDX05380.1 KQ435022 KZC13921.1 CCAG010022043 CH480822 EDW55631.1 CM002910 KMY90823.1 KQ982877 KYQ49709.1 CH954179 EDV54780.1 KQ980662 KYN14715.1 QOIP01000010 RLU17910.1 JXJN01008376 KRF81957.1 KB632075 ERL88558.1 NEVH01003493 PNF40733.1 KPU73076.1 KPU73077.1 KPU73078.1 KQ435912 KOX68884.1 GAPW01000041 JAC13557.1 KQ978311 KYM95318.1 GL763048 EFZ20307.1 APGK01050727 APGK01050728 APGK01050729 APGK01050730 APGK01050731 APGK01050732 APGK01050733 APGK01050734 APGK01050735 KB741176 ENN73201.1 GL442519 EFN63376.1 GGFK01008426 MBW41747.1 KQ414665 KOC65152.1 GAPW01000061 JAC13537.1 PYGN01001063 PSN37939.1 LJIG01009350 KRT83256.1 DS235170 EEB12828.1 UFQT01000026 SSX18052.1 SSX18053.1 GAPW01000082 JAC13516.1 GDHC01000678 JAQ17951.1 GBHO01044318 GBHO01037253 JAF99285.1 JAG06351.1
Proteomes
UP000005204
UP000053240
UP000283053
UP000053268
UP000007151
UP000218220
+ More
UP000075900 UP000075885 UP000076408 UP000075886 UP000075880 UP000075882 UP000075884 UP000007062 UP000075903 UP000030765 UP000075883 UP000183832 UP000075881 UP000075920 UP000002320 UP000007266 UP000069940 UP000249989 UP000008820 UP000075840 UP000076407 UP000192223 UP000007798 UP000095301 UP000009192 UP000078200 UP000092553 UP000008792 UP000001819 UP000095300 UP000268350 UP000037069 UP000007801 UP000001070 UP000092462 UP000005205 UP000008237 UP000078540 UP000007755 UP000192221 UP000002282 UP000000803 UP000078541 UP000000304 UP000076502 UP000092444 UP000001292 UP000092445 UP000075809 UP000008711 UP000078492 UP000279307 UP000092460 UP000092443 UP000005203 UP000030742 UP000235965 UP000053105 UP000091820 UP000078542 UP000019118 UP000000311 UP000053825 UP000245037 UP000009046
UP000075900 UP000075885 UP000076408 UP000075886 UP000075880 UP000075882 UP000075884 UP000007062 UP000075903 UP000030765 UP000075883 UP000183832 UP000075881 UP000075920 UP000002320 UP000007266 UP000069940 UP000249989 UP000008820 UP000075840 UP000076407 UP000192223 UP000007798 UP000095301 UP000009192 UP000078200 UP000092553 UP000008792 UP000001819 UP000095300 UP000268350 UP000037069 UP000007801 UP000001070 UP000092462 UP000005205 UP000008237 UP000078540 UP000007755 UP000192221 UP000002282 UP000000803 UP000078541 UP000000304 UP000076502 UP000092444 UP000001292 UP000092445 UP000075809 UP000008711 UP000078492 UP000279307 UP000092460 UP000092443 UP000005203 UP000030742 UP000235965 UP000053105 UP000091820 UP000078542 UP000019118 UP000000311 UP000053825 UP000245037 UP000009046
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JBZ7
A0A2H1V903
A0A0N1IPH7
A0A3S2LDE4
A0A194PPD9
A0A212FP75
+ More
A0A2A4J6A6 A0A1Y1K115 A0A182RDN6 A0A182PUE0 A0A182Y399 A0A182QXR0 A0A182JK69 A0A182L0N8 A0A182NTX0 Q7Q4D6 A0A1S4GY16 A0A182UMG8 A0A2M3YZ06 A0A2M3YZ58 A0A2M4B9C9 A0A084W5U2 A0A2M4CTY0 A0A182LTI9 A0A1J1I1B5 A0A2M4CTF6 A0A2M4CTI2 A0A182K1Q9 A0A182W6I3 B0WXB4 D6WXZ9 A0A023EWS4 A0A182H8D0 Q0IFX2 A0A182IC95 A0A182XLC5 A0A1W4XF82 B4MZG3 A0A1I8N7R6 T1PE43 B4KK29 A0A1A9VXM6 A0A034VC57 A0A0M4E6Z3 A0A0K8VET0 A0A0A1XK54 B4LTJ0 Q29MP6 A0A0A1WM92 A0A1I8P8B2 A0A3B0K071 A0A0L0CBR6 B3MM24 A0A310SLN5 B4JDM1 A0A1B0DMT2 A0A158NFQ1 E2C4H1 A0A195BDL5 F4WGS9 A0A1W4V9M9 B4P9U9 Q9VJ80 A0A151JVI7 B4Q8U8 A0A154PPW0 A0A1B0FN29 B4I5H5 A0A1A9ZIV4 A0A0J9R404 A0A151WP86 B3NMA9 A0A195DP72 A0A3L8DBM2 A0A1B0B4N1 A0A1A9XDR7 A0A087ZYF6 A0A0Q9WNC9 U4U893 A0A2J7RIR8 A0A0P9BMS4 A0A0N0BCF0 A0A1A9W0L4 A0A023EWT3 A0A151I936 E9IGL4 N6TXU1 E2AT66 A0A2M4ALX6 A0A0L7R2Q0 A0A023EWS2 A0A2P8Y122 A0A0T6B7C2 E0VHH2 A0A336LIY5 A0A336LN33 A0A023EX84 A0A146MDQ2 A0A0A9VVR2
A0A2A4J6A6 A0A1Y1K115 A0A182RDN6 A0A182PUE0 A0A182Y399 A0A182QXR0 A0A182JK69 A0A182L0N8 A0A182NTX0 Q7Q4D6 A0A1S4GY16 A0A182UMG8 A0A2M3YZ06 A0A2M3YZ58 A0A2M4B9C9 A0A084W5U2 A0A2M4CTY0 A0A182LTI9 A0A1J1I1B5 A0A2M4CTF6 A0A2M4CTI2 A0A182K1Q9 A0A182W6I3 B0WXB4 D6WXZ9 A0A023EWS4 A0A182H8D0 Q0IFX2 A0A182IC95 A0A182XLC5 A0A1W4XF82 B4MZG3 A0A1I8N7R6 T1PE43 B4KK29 A0A1A9VXM6 A0A034VC57 A0A0M4E6Z3 A0A0K8VET0 A0A0A1XK54 B4LTJ0 Q29MP6 A0A0A1WM92 A0A1I8P8B2 A0A3B0K071 A0A0L0CBR6 B3MM24 A0A310SLN5 B4JDM1 A0A1B0DMT2 A0A158NFQ1 E2C4H1 A0A195BDL5 F4WGS9 A0A1W4V9M9 B4P9U9 Q9VJ80 A0A151JVI7 B4Q8U8 A0A154PPW0 A0A1B0FN29 B4I5H5 A0A1A9ZIV4 A0A0J9R404 A0A151WP86 B3NMA9 A0A195DP72 A0A3L8DBM2 A0A1B0B4N1 A0A1A9XDR7 A0A087ZYF6 A0A0Q9WNC9 U4U893 A0A2J7RIR8 A0A0P9BMS4 A0A0N0BCF0 A0A1A9W0L4 A0A023EWT3 A0A151I936 E9IGL4 N6TXU1 E2AT66 A0A2M4ALX6 A0A0L7R2Q0 A0A023EWS2 A0A2P8Y122 A0A0T6B7C2 E0VHH2 A0A336LIY5 A0A336LN33 A0A023EX84 A0A146MDQ2 A0A0A9VVR2
PDB
6BMT
E-value=4.05451e-87,
Score=824
Ontologies
GO
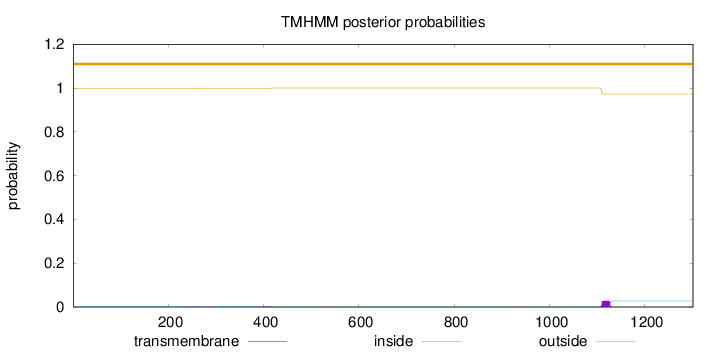
Topology
Length:
1301
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.55727
Exp number, first 60 AAs:
2e-05
Total prob of N-in:
0.00128
outside
1 - 1301
Population Genetic Test Statistics
Pi
272.619306
Theta
207.571478
Tajima's D
0.138341
CLR
0.49911
CSRT
0.406129693515324
Interpretation
Uncertain
