Gene
KWMTBOMO10381 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007043
Annotation
PREDICTED:_signal_peptidase_complex_subunit_1_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 1.967
Sequence
CDS
ATGGATTTCTTTACATCTATACCAACCCACATAGATTACGTCGGACAAGCAAAAGCAGAGAAATTATACAGGGCAATAATTACATTGTTCAGTATAGTTGGTTTTGTATGGGGTTATATAGTTCAGCAATTCTCGCAGTCAGTGTATATTCTTGGTGCTGGATTTCTGCTTGCTGCCATTCTTACTGTTCCTCCATGGCCAATGTACCGACGCAATCCTCTGAACTGGCAGGTTCCTAAGAACGGTGATGACAAACCAGCAGCCAAGAAAGGAAAGAAGTGA
Protein
MDFFTSIPTHIDYVGQAKAEKLYRAIITLFSIVGFVWGYIVQQFSQSVYILGAGFLLAAILTVPPWPMYRRNPLNWQVPKNGDDKPAAKKGKK
Summary
Similarity
Belongs to the SLC26A/SulP transporter (TC 2.A.53) family.
Uniprot
H9JBZ8
A0A2H1V8V7
A0A0N1IPQ6
A0A0L7L175
I4DMW5
I4DJT5
+ More
A0A212FPB9 S4NSY9 A0A1W4WTR3 A0A151X5E0 R4FPD3 E2ABG7 A0A023FB15 E2J7F1 A0A069DN76 T1HXR1 A0A158NAG5 A0A0J7KM77 G1K074 A0A195CKT1 D6WSR1 A0A232FGC7 K7J753 E2BXX4 A0A026WH36 E0VNM3 A0A0P4VVC5 A0A067QGU3 A0A0L7R8H6 C1BNH2 A0A0N0BIL7 C1C1A0 A0A0C9R5S7 V9IMD8 A0A088A5E0 C1BSI6 A0A154NZ69 A0A0L0BZ22 A0A154P107 W4ZAU7 A0A0K2SWX1 A0A023EEJ8 A0A182GPY6 A0A023EE46 Q17C90 A0A1S4F884 A0A182PRH3 A0A182Q499 A0A182R5M3 Q1HR98 A0A182VY52 A0A182X8V0 A0A182VLU2 A0A182L6W9 Q7QAD1 A0A336LSM9 A0A182N4Q3 A0A182XWQ7 T1E3W6 A0A0K8TPL6 A0A1Q3FGP7 A0A084VEN7 A0A1I8M0G9 A0A182F9K6 A0A2M4A8P8 W5JCK6 A0A0N0BIG4 U5ELA0 A0A2M3ZL96 A0A2M4C4P1 A0A1I8NYG0 A0A182IZC6 A0A1L8EFC0 A0A1L8EFH2 A0A1L8EFE7 N6TQ36 B0WEY3 C4N1A3 A0A1B0B519 A0A3M6T9R9 B4KCJ5 A0A151J0H9 A0A2J7RFH4 A0A1D2MQ40 A0A1S3IXF4 A0A0M4ENC6 A0A195EVM6 A0A1Q3FH21 Q29BW7 B4GPK2 T1J4D0 A0A1B0EUG8 A0A3Q2YTG4 R7UNQ8 A0A3Q2E0F5 A0A3Q3KB63 A0A1I8P5K5 A0A3P9N4F0 T1KH60 A0A3B3YWQ5
A0A212FPB9 S4NSY9 A0A1W4WTR3 A0A151X5E0 R4FPD3 E2ABG7 A0A023FB15 E2J7F1 A0A069DN76 T1HXR1 A0A158NAG5 A0A0J7KM77 G1K074 A0A195CKT1 D6WSR1 A0A232FGC7 K7J753 E2BXX4 A0A026WH36 E0VNM3 A0A0P4VVC5 A0A067QGU3 A0A0L7R8H6 C1BNH2 A0A0N0BIL7 C1C1A0 A0A0C9R5S7 V9IMD8 A0A088A5E0 C1BSI6 A0A154NZ69 A0A0L0BZ22 A0A154P107 W4ZAU7 A0A0K2SWX1 A0A023EEJ8 A0A182GPY6 A0A023EE46 Q17C90 A0A1S4F884 A0A182PRH3 A0A182Q499 A0A182R5M3 Q1HR98 A0A182VY52 A0A182X8V0 A0A182VLU2 A0A182L6W9 Q7QAD1 A0A336LSM9 A0A182N4Q3 A0A182XWQ7 T1E3W6 A0A0K8TPL6 A0A1Q3FGP7 A0A084VEN7 A0A1I8M0G9 A0A182F9K6 A0A2M4A8P8 W5JCK6 A0A0N0BIG4 U5ELA0 A0A2M3ZL96 A0A2M4C4P1 A0A1I8NYG0 A0A182IZC6 A0A1L8EFC0 A0A1L8EFH2 A0A1L8EFE7 N6TQ36 B0WEY3 C4N1A3 A0A1B0B519 A0A3M6T9R9 B4KCJ5 A0A151J0H9 A0A2J7RFH4 A0A1D2MQ40 A0A1S3IXF4 A0A0M4ENC6 A0A195EVM6 A0A1Q3FH21 Q29BW7 B4GPK2 T1J4D0 A0A1B0EUG8 A0A3Q2YTG4 R7UNQ8 A0A3Q2E0F5 A0A3Q3KB63 A0A1I8P5K5 A0A3P9N4F0 T1KH60 A0A3B3YWQ5
Pubmed
19121390
26354079
26227816
22651552
22118469
23622113
+ More
20798317 25474469 26334808 21347285 21736942 18362917 19820115 28648823 20075255 24508170 30249741 20566863 27129103 24845553 26108605 24945155 26483478 17510324 17204158 20966253 12364791 14747013 17210077 25244985 24330624 26369729 24438588 25315136 20920257 23761445 23537049 19576987 30382153 17994087 27289101 15632085 23254933
20798317 25474469 26334808 21347285 21736942 18362917 19820115 28648823 20075255 24508170 30249741 20566863 27129103 24845553 26108605 24945155 26483478 17510324 17204158 20966253 12364791 14747013 17210077 25244985 24330624 26369729 24438588 25315136 20920257 23761445 23537049 19576987 30382153 17994087 27289101 15632085 23254933
EMBL
BABH01014715
ODYU01001281
SOQ37283.1
KQ460328
KPJ15659.1
JTDY01003687
+ More
KOB69150.1 AK402633 BAM19255.1 AK401553 KQ459603 BAM18175.1 KPI93002.1 AGBW02003464 OWR55550.1 GAIX01013852 JAA78708.1 KQ982519 KYQ55509.1 GAHY01000939 JAA76571.1 GL438246 EFN69233.1 GBBI01000006 JAC18706.1 HP429369 ADN29869.1 GBGD01003609 JAC85280.1 ACPB03017388 ADTU01010286 LBMM01005592 KMQ91397.1 JO494955 AEL79185.1 KQ977642 KYN01062.1 KQ971354 EFA07146.1 NNAY01000297 OXU29377.1 AAZX01008222 GL451341 EFN79457.1 KK107231 QOIP01000009 EZA54986.1 RLU19097.1 DS235340 EEB14979.1 GDKW01003032 JAI53563.1 KK853474 KDR07356.1 KQ414633 KOC67066.1 BT076151 BT076647 BT077217 ACO10575.1 ACO11071.1 KQ435729 KOX77641.1 BT080629 ACO15053.1 GBYB01003280 JAG73047.1 JR051784 AEY61559.1 BT077565 ACO11989.1 KQ434783 KZC04927.1 JRES01001133 KNC25260.1 KZC04928.1 AAGJ04052850 HACA01000932 CDW18293.1 GAPW01006519 JAC07079.1 JXUM01015410 KQ560429 KXJ82450.1 GAPW01006518 GAPW01006517 JAC07081.1 CH477310 EAT43985.1 AXCN02001188 DQ440196 ABF18229.1 AAAB01008898 EAA09008.3 UFQS01000078 UFQT01000078 SSW99045.1 SSX19427.1 GALA01000005 JAA94847.1 GDAI01001299 JAI16304.1 GFDL01008324 JAV26721.1 ATLV01012285 KE524778 KFB36431.1 GGFK01003791 MBW37112.1 ADMH02001608 ETN61821.1 KOX77642.1 GANO01001496 JAB58375.1 GGFM01008437 MBW29188.1 GGFJ01011074 MBW60215.1 GFDG01001382 JAV17417.1 GFDG01001385 JAV17414.1 GFDG01001383 JAV17416.1 APGK01025593 KB740597 ENN80128.1 DS231912 EDS25841.1 EZ048876 ACN69168.1 JXJN01008590 RCHS01004051 RMX38081.1 CH933806 EDW14814.1 KQ980629 KYN15016.1 NEVH01004413 PNF39577.1 LJIJ01000696 ODM95220.1 CP012526 ALC46176.1 KQ981954 KYN32196.1 GFDL01008211 JAV26834.1 CM000070 EAL26879.1 CH479186 EDW39085.1 JH431842 AJWK01015109 AMQN01007790 KB301402 ELU05542.1 CAEY01000073
KOB69150.1 AK402633 BAM19255.1 AK401553 KQ459603 BAM18175.1 KPI93002.1 AGBW02003464 OWR55550.1 GAIX01013852 JAA78708.1 KQ982519 KYQ55509.1 GAHY01000939 JAA76571.1 GL438246 EFN69233.1 GBBI01000006 JAC18706.1 HP429369 ADN29869.1 GBGD01003609 JAC85280.1 ACPB03017388 ADTU01010286 LBMM01005592 KMQ91397.1 JO494955 AEL79185.1 KQ977642 KYN01062.1 KQ971354 EFA07146.1 NNAY01000297 OXU29377.1 AAZX01008222 GL451341 EFN79457.1 KK107231 QOIP01000009 EZA54986.1 RLU19097.1 DS235340 EEB14979.1 GDKW01003032 JAI53563.1 KK853474 KDR07356.1 KQ414633 KOC67066.1 BT076151 BT076647 BT077217 ACO10575.1 ACO11071.1 KQ435729 KOX77641.1 BT080629 ACO15053.1 GBYB01003280 JAG73047.1 JR051784 AEY61559.1 BT077565 ACO11989.1 KQ434783 KZC04927.1 JRES01001133 KNC25260.1 KZC04928.1 AAGJ04052850 HACA01000932 CDW18293.1 GAPW01006519 JAC07079.1 JXUM01015410 KQ560429 KXJ82450.1 GAPW01006518 GAPW01006517 JAC07081.1 CH477310 EAT43985.1 AXCN02001188 DQ440196 ABF18229.1 AAAB01008898 EAA09008.3 UFQS01000078 UFQT01000078 SSW99045.1 SSX19427.1 GALA01000005 JAA94847.1 GDAI01001299 JAI16304.1 GFDL01008324 JAV26721.1 ATLV01012285 KE524778 KFB36431.1 GGFK01003791 MBW37112.1 ADMH02001608 ETN61821.1 KOX77642.1 GANO01001496 JAB58375.1 GGFM01008437 MBW29188.1 GGFJ01011074 MBW60215.1 GFDG01001382 JAV17417.1 GFDG01001385 JAV17414.1 GFDG01001383 JAV17416.1 APGK01025593 KB740597 ENN80128.1 DS231912 EDS25841.1 EZ048876 ACN69168.1 JXJN01008590 RCHS01004051 RMX38081.1 CH933806 EDW14814.1 KQ980629 KYN15016.1 NEVH01004413 PNF39577.1 LJIJ01000696 ODM95220.1 CP012526 ALC46176.1 KQ981954 KYN32196.1 GFDL01008211 JAV26834.1 CM000070 EAL26879.1 CH479186 EDW39085.1 JH431842 AJWK01015109 AMQN01007790 KB301402 ELU05542.1 CAEY01000073
Proteomes
UP000005204
UP000053240
UP000037510
UP000053268
UP000007151
UP000192223
+ More
UP000075809 UP000000311 UP000015103 UP000005205 UP000036403 UP000078542 UP000007266 UP000215335 UP000002358 UP000008237 UP000053097 UP000279307 UP000009046 UP000027135 UP000053825 UP000053105 UP000005203 UP000076502 UP000037069 UP000007110 UP000069940 UP000249989 UP000008820 UP000075885 UP000075886 UP000075900 UP000075920 UP000076407 UP000075903 UP000075882 UP000007062 UP000075884 UP000076408 UP000030765 UP000095301 UP000069272 UP000000673 UP000095300 UP000075880 UP000019118 UP000002320 UP000092460 UP000275408 UP000009192 UP000078492 UP000235965 UP000094527 UP000085678 UP000092553 UP000078541 UP000001819 UP000008744 UP000092461 UP000264820 UP000014760 UP000265020 UP000261600 UP000242638 UP000015104 UP000261480
UP000075809 UP000000311 UP000015103 UP000005205 UP000036403 UP000078542 UP000007266 UP000215335 UP000002358 UP000008237 UP000053097 UP000279307 UP000009046 UP000027135 UP000053825 UP000053105 UP000005203 UP000076502 UP000037069 UP000007110 UP000069940 UP000249989 UP000008820 UP000075885 UP000075886 UP000075900 UP000075920 UP000076407 UP000075903 UP000075882 UP000007062 UP000075884 UP000076408 UP000030765 UP000095301 UP000069272 UP000000673 UP000095300 UP000075880 UP000019118 UP000002320 UP000092460 UP000275408 UP000009192 UP000078492 UP000235965 UP000094527 UP000085678 UP000092553 UP000078541 UP000001819 UP000008744 UP000092461 UP000264820 UP000014760 UP000265020 UP000261600 UP000242638 UP000015104 UP000261480
PRIDE
Pfam
Interpro
IPR009542
SPC12
+ More
IPR037713 Spc1
IPR001902 SLC26A/SulP_fam
IPR002645 STAS_dom
IPR011547 SLC26A/SulP_dom
IPR036513 STAS_dom_sf
IPR013320 ConA-like_dom_sf
IPR026847 VPS13
IPR041969 VP13D_UBA
IPR001870 B30.2/SPRY
IPR009543 SHR-BD
IPR015940 UBA
IPR001496 SOCS_box
IPR026854 VPS13_N
IPR000772 Ricin_B_lectin
IPR003877 SPRY_dom
IPR009060 UBA-like_sf
IPR031645 VPS13_C
IPR031642 VPS13_mid_rpt
IPR031646 VPS13_N2
IPR037713 Spc1
IPR001902 SLC26A/SulP_fam
IPR002645 STAS_dom
IPR011547 SLC26A/SulP_dom
IPR036513 STAS_dom_sf
IPR013320 ConA-like_dom_sf
IPR026847 VPS13
IPR041969 VP13D_UBA
IPR001870 B30.2/SPRY
IPR009543 SHR-BD
IPR015940 UBA
IPR001496 SOCS_box
IPR026854 VPS13_N
IPR000772 Ricin_B_lectin
IPR003877 SPRY_dom
IPR009060 UBA-like_sf
IPR031645 VPS13_C
IPR031642 VPS13_mid_rpt
IPR031646 VPS13_N2
Gene 3D
ProteinModelPortal
H9JBZ8
A0A2H1V8V7
A0A0N1IPQ6
A0A0L7L175
I4DMW5
I4DJT5
+ More
A0A212FPB9 S4NSY9 A0A1W4WTR3 A0A151X5E0 R4FPD3 E2ABG7 A0A023FB15 E2J7F1 A0A069DN76 T1HXR1 A0A158NAG5 A0A0J7KM77 G1K074 A0A195CKT1 D6WSR1 A0A232FGC7 K7J753 E2BXX4 A0A026WH36 E0VNM3 A0A0P4VVC5 A0A067QGU3 A0A0L7R8H6 C1BNH2 A0A0N0BIL7 C1C1A0 A0A0C9R5S7 V9IMD8 A0A088A5E0 C1BSI6 A0A154NZ69 A0A0L0BZ22 A0A154P107 W4ZAU7 A0A0K2SWX1 A0A023EEJ8 A0A182GPY6 A0A023EE46 Q17C90 A0A1S4F884 A0A182PRH3 A0A182Q499 A0A182R5M3 Q1HR98 A0A182VY52 A0A182X8V0 A0A182VLU2 A0A182L6W9 Q7QAD1 A0A336LSM9 A0A182N4Q3 A0A182XWQ7 T1E3W6 A0A0K8TPL6 A0A1Q3FGP7 A0A084VEN7 A0A1I8M0G9 A0A182F9K6 A0A2M4A8P8 W5JCK6 A0A0N0BIG4 U5ELA0 A0A2M3ZL96 A0A2M4C4P1 A0A1I8NYG0 A0A182IZC6 A0A1L8EFC0 A0A1L8EFH2 A0A1L8EFE7 N6TQ36 B0WEY3 C4N1A3 A0A1B0B519 A0A3M6T9R9 B4KCJ5 A0A151J0H9 A0A2J7RFH4 A0A1D2MQ40 A0A1S3IXF4 A0A0M4ENC6 A0A195EVM6 A0A1Q3FH21 Q29BW7 B4GPK2 T1J4D0 A0A1B0EUG8 A0A3Q2YTG4 R7UNQ8 A0A3Q2E0F5 A0A3Q3KB63 A0A1I8P5K5 A0A3P9N4F0 T1KH60 A0A3B3YWQ5
A0A212FPB9 S4NSY9 A0A1W4WTR3 A0A151X5E0 R4FPD3 E2ABG7 A0A023FB15 E2J7F1 A0A069DN76 T1HXR1 A0A158NAG5 A0A0J7KM77 G1K074 A0A195CKT1 D6WSR1 A0A232FGC7 K7J753 E2BXX4 A0A026WH36 E0VNM3 A0A0P4VVC5 A0A067QGU3 A0A0L7R8H6 C1BNH2 A0A0N0BIL7 C1C1A0 A0A0C9R5S7 V9IMD8 A0A088A5E0 C1BSI6 A0A154NZ69 A0A0L0BZ22 A0A154P107 W4ZAU7 A0A0K2SWX1 A0A023EEJ8 A0A182GPY6 A0A023EE46 Q17C90 A0A1S4F884 A0A182PRH3 A0A182Q499 A0A182R5M3 Q1HR98 A0A182VY52 A0A182X8V0 A0A182VLU2 A0A182L6W9 Q7QAD1 A0A336LSM9 A0A182N4Q3 A0A182XWQ7 T1E3W6 A0A0K8TPL6 A0A1Q3FGP7 A0A084VEN7 A0A1I8M0G9 A0A182F9K6 A0A2M4A8P8 W5JCK6 A0A0N0BIG4 U5ELA0 A0A2M3ZL96 A0A2M4C4P1 A0A1I8NYG0 A0A182IZC6 A0A1L8EFC0 A0A1L8EFH2 A0A1L8EFE7 N6TQ36 B0WEY3 C4N1A3 A0A1B0B519 A0A3M6T9R9 B4KCJ5 A0A151J0H9 A0A2J7RFH4 A0A1D2MQ40 A0A1S3IXF4 A0A0M4ENC6 A0A195EVM6 A0A1Q3FH21 Q29BW7 B4GPK2 T1J4D0 A0A1B0EUG8 A0A3Q2YTG4 R7UNQ8 A0A3Q2E0F5 A0A3Q3KB63 A0A1I8P5K5 A0A3P9N4F0 T1KH60 A0A3B3YWQ5
Ontologies
GO
PANTHER
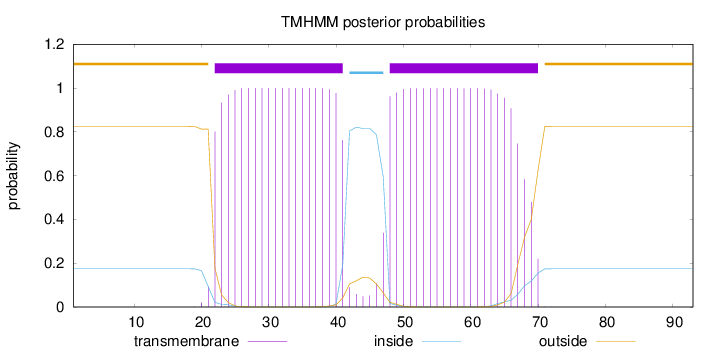
Topology
Subcellular location
Membrane
Length:
93
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
41.01627
Exp number, first 60 AAs:
33.15721
Total prob of N-in:
0.17459
POSSIBLE N-term signal
sequence
outside
1 - 21
TMhelix
22 - 41
inside
42 - 47
TMhelix
48 - 70
outside
71 - 93
Population Genetic Test Statistics
Pi
232.054562
Theta
182.050155
Tajima's D
0.807622
CLR
0
CSRT
0.605969701514924
Interpretation
Uncertain
