Gene
KWMTBOMO10380
Pre Gene Modal
BGIBMGA007063
Annotation
PREDICTED:_GPI_inositol-deacylase_[Amyelois_transitella]
Full name
GPI inositol-deacylase
Location in the cell
PlasmaMembrane Reliability : 4.446
Sequence
CDS
ATGTTTGAATATCCACAATTTGTGCATCTGTCTATGCCAGAAAACAAAATTTATCCACAATATGCCTTATTTGCATATAGTGAAGGCAGATTCACAGAGAGAGCAAGAAAAATGTGGTTTGACGGAGTACCAGTGTTATTCATACCAGGCAATTCTGGTAGTCACATGCAAGCAAGATCACTGGCATCTGTGGCCTTAAGAAAAGCCTTGTCTGGAGGATATGATTACCATTTTGATTACTTTGCCATAAGCTATAATGAGGAGCTCTCAGGCCTTTATGGAGGAGTTCTACAAAGTCAAACAAGATTTGCAGCTGCTTGTATAGAAAAAATATTGGATCTGTATAAAGAAAATAATTATATTAAATCTGTACCAACATCTGTTATCCTCATTGGACATTCAATGGGAGGTCTTGTAGCCAAGCGTTTGTTGGTTCATCCAAGTACAGTCAATTCGACAAGTATAGCAATCACTTTAGCAGCTCCTTTAGAAGCCCCACTGATTAACTTTGATATAGCTATGAATGATTACTATTTAAGTATGAATCATGAATGGGAAAATTTCGTAAGAGGCAATCAATTGATGACGGATAAGAAGATGCTTTTAAGTTTTGGAAATGGACCAAGAGATTTACTAATGCCTTCAGGCTTAACATTATCAAACGACAGTGATATAAATACATTGACAACAGCGATACCAGGCGTCTGGGTATCACCAGATCACGTCTGTATTGTATGGTGCAAGCAACTTGTTATGGCAATAAACAAATACCTATTTTCGATAGTTAATCCAATGACGAAACAAATAAGTTATGATAAACAGTTGCTAATGTCGAGAGCCAAGCAGTATTTTCAGGCGAATCGTTCAATGACAATTAATCCAGAAATTCCTAAAGCCAATGTGACTATGACAGCGGATGCGTTCTGGTACGAAGATAACCGGAGGATGTATCAAGTGCAACGACCGGAAATTGACAAAACAACATATCTAATGATAAGATTAGTTAAATTCCCGCAGAACAGATTCGTAGCTGTCGAGGCTGTTAATGTGTCTGATAAAGATTGGATATATGGATGTAACGCGAAACATACATACAACACGCACAGATATTGTAAACACGCAGTATCGTTAACAGAATTAAGTCGTTGGTCCGGAGCTGCGACCGAATTTCGAAAGAGGAAGTTAGCAACAGTGAACCTGCACAATGTGATGGAACATTACCCGGATTGGTCTCATGTTGTCGTCAAGGTATCGCCTACACGAAAACCCGTAACTATGAACATCGACATCAACGATTATGCGTCCCGGCAGATATTCGTGGATCTACCCACGTTTTTTGAATTCGGAAAGAGGATCGTGAAACCAGAAACCGAACGAGACAGTTTGTATTACGAATTGATCTTGCGAGATTTTAATGCGATACACGAAGCGTACCTCCTTTACGTTGAGCCGACTGCGAATTGTAAAGCAAGCCGATATCACGTTTCGGCGGAGTTGCATGTTCCGTGGGCGAAAAATCACGAATATTATCATTATTTTACGCATTTGAAAAGGTCGCCTATGAAGTTAAGGCTCTTTAAAACAAACCCTAATTTAACACTTGGGTTGGAAACCACCGAGCACGCGAAAATTACTTTGCTATTGGATCCTCAATGTCAATTTATTCTCAGTATATCGTCATCGTGGTACAATCGCCTCGCTCAGTTTACTCGAAATTACGGTCCAGTACTTATTCCGTATGTAGCCGCCATAGTGTTATTAGCTGCCAGAAGTATTATTATGAACTTTCAAGAATCTGGATCATGCTTGACCATTCATGGAGCTTTAATGAGCGAAGGCGTCAAACCCTATTATTGTCTCGTGTTTTCAAGATTAGCAACTAAGATTTTTTTGGCGATTCCGCTGTTCTCATTTATGTTCGAAAGCGCAAGCTGGAGGGATCTCGAATTGCAGTATTTTATCAAATCCCTGTTAGTGTTGCCAGCTTATATGACAGCATTAGGGATTGTCAATATTGTAGCTGCTGCTATAATGGGTGCAATGGTGTTTTCATCACAGATAGCGCATAGATTACTGTTTAAAATAGTATGGCGAGGCGGTGGTAGTTTAGCAGAGAAAATGGCCACCGGATTGCACAAGCTCCCCATGATCGTGAGTGCAGCGCTAGTTTGTGCGGTTCCTTTATCATGCGGAGCAGCTTCATTGGCTGCCGGTGCCGCATTTTATGCATTTATGCTCTCGAAAATGTATGAAGAATATTTAGAAGATTACGTTTACAAAATAATGGCAAAGCTAGCTTCTAAAGTCTGTCGTATGTTCAACTCGACAAGAAACAAATATGAAAAATCTGAAACCAATTTGATTTCAAAAGCTTTATCGGATGCAAAGGAAAGTAATCTCAGATCTAAAGAAATTGACGAAATTGAAGGTAATGATAAGGATTCAACGAACAATGTAGGACAAGACGGTTCTTCAAACACAGACTGTGCTTTAGTTTCTGCGAATAAAGAAAGCGAACGAAAGCCCAAAGAAGGCAGTATTAAAGGTGAAAGCAAAGACTTGGACGAGGATTTTAATAGCATTAACTTCCACATGATGTTGTTTTTCATGTGGATCACAGTAACTCTAGTCAATGTTCCAGCATTATTAACATGGGCGCGAAACTTTAAGTATTCCATGGTACTAAAACCAGATACGTCATATCACACTGGTCTAGTCATGTCAGTTTGTTCCAGTTGTATCTGGCAAATCAATGGTCCCAGAAGAAATTTGAAGCATTATGAAGCGGTTTCATCTTTACTATTTACAATGGCTATATTTATATTAGCTTTGGGACCCCTATCACTCACAATAGTCAACTATGGAATCACTTTCACATTTGTCGTTATTACATTACAACAAGTATGTGATAAGGAAGATGTTCAAAGTTCTACCATTGAAGGTACAAATGAATCCAAGAATGATAATGATGAAAAACCAAAAGAAAACCAAAAAGAAGAAGCTGATAATATTCAAACGAATGATTGTGATCCATGCAATGAGAACAGAATATTTGATGCTTTTAAGAATCTTCAAGATAAATTTGATTTAACTACAATGGATAATTTGTAA
Protein
MFEYPQFVHLSMPENKIYPQYALFAYSEGRFTERARKMWFDGVPVLFIPGNSGSHMQARSLASVALRKALSGGYDYHFDYFAISYNEELSGLYGGVLQSQTRFAAACIEKILDLYKENNYIKSVPTSVILIGHSMGGLVAKRLLVHPSTVNSTSIAITLAAPLEAPLINFDIAMNDYYLSMNHEWENFVRGNQLMTDKKMLLSFGNGPRDLLMPSGLTLSNDSDINTLTTAIPGVWVSPDHVCIVWCKQLVMAINKYLFSIVNPMTKQISYDKQLLMSRAKQYFQANRSMTINPEIPKANVTMTADAFWYEDNRRMYQVQRPEIDKTTYLMIRLVKFPQNRFVAVEAVNVSDKDWIYGCNAKHTYNTHRYCKHAVSLTELSRWSGAATEFRKRKLATVNLHNVMEHYPDWSHVVVKVSPTRKPVTMNIDINDYASRQIFVDLPTFFEFGKRIVKPETERDSLYYELILRDFNAIHEAYLLYVEPTANCKASRYHVSAELHVPWAKNHEYYHYFTHLKRSPMKLRLFKTNPNLTLGLETTEHAKITLLLDPQCQFILSISSSWYNRLAQFTRNYGPVLIPYVAAIVLLAARSIIMNFQESGSCLTIHGALMSEGVKPYYCLVFSRLATKIFLAIPLFSFMFESASWRDLELQYFIKSLLVLPAYMTALGIVNIVAAAIMGAMVFSSQIAHRLLFKIVWRGGGSLAEKMATGLHKLPMIVSAALVCAVPLSCGAASLAAGAAFYAFMLSKMYEEYLEDYVYKIMAKLASKVCRMFNSTRNKYEKSETNLISKALSDAKESNLRSKEIDEIEGNDKDSTNNVGQDGSSNTDCALVSANKESERKPKEGSIKGESKDLDEDFNSINFHMMLFFMWITVTLVNVPALLTWARNFKYSMVLKPDTSYHTGLVMSVCSSCIWQINGPRRNLKHYEAVSSLLFTMAIFILALGPLSLTIVNYGITFTFVVITLQQVCDKEDVQSSTIEGTNESKNDNDEKPKENQKEEADNIQTNDCDPCNENRIFDAFKNLQDKFDLTTMDNL
Summary
Description
Involved in inositol deacylation of GPI-anchored proteins which plays important roles in the quality control and ER-associated degradation of GPI-anchored proteins.
Involved in inositol deacylation of GPI-anchored proteins. GPI inositol deacylation may important for efficient transport of GPI-anchored proteins from the endoplasmic reticulum to the Golgi.
Involved in inositol deacylation of GPI-anchored proteins. GPI inositol deacylation may important for efficient transport of GPI-anchored proteins from the endoplasmic reticulum to the Golgi.
Similarity
Belongs to the GPI inositol-deacylase family.
Feature
chain GPI inositol-deacylase
Uniprot
A0A212FP83
A0A2H1VAP4
A0A0L7L106
A0A067QZ37
A0A1W4WDY2
U5EZS7
+ More
A0A2J7R6B4 A0A336LRH5 A0A1J1HJN1 A0A1Y1N9E7 A0A1L8DUQ7 A0A1Q3G1I1 A0A1Q3G1J6 D6WW73 A0A1S4FY25 Q16K53 A0A182G874 A0A2P8XLK3 A0A182QD08 E0VHG5 A0A2M4BCT8 A0A2M4BCA0 A0A084VXD6 Q7PSQ5 V5GS79 W5JN50 A0A2M4AGP6 A0A182FLK3 A0A2M3Z242 A0A2M4AGR0 A0A2M3Z263 A0A146KTB8 A0A1I8N7F8 A0A182J144 A0A210QVH4 B4M812 A0A3B0KSD9 A0A1Y1N5V3 A0A0M9A5B9 B4H4F9 Q29I09 A0A088A5S0 W8BQ10 A0A0M4EP13 A0A3R7PVS9 B4JKJ5 A0A0P5Y5F1 A0A0P4Z516 A0A0P5B899 A0A0P5V869 U3JNW4 A0A0P5Q8S3 A0A0N8CCQ3 A0A0N8C2A5 A0A164K7R0 A0A0P6G1F3 A0A0P6HHW7 A0A0P6G852 V9K8G2 A0A1I8PZ32 A0A0P6CUV9 A0A0N8DVT0 A0A0K8U5B9 A0A0P4Z553 A0A0P4YXQ4 A0A0K8V6V2 A0A0P5Y2F6 A0A0P5N884 C3YRU1 A0A0A0AV20 A0A091VBE1 A0A026WKA5 A0A151WLQ7 A0A0P5IVF2 A0A2I0U480
A0A2J7R6B4 A0A336LRH5 A0A1J1HJN1 A0A1Y1N9E7 A0A1L8DUQ7 A0A1Q3G1I1 A0A1Q3G1J6 D6WW73 A0A1S4FY25 Q16K53 A0A182G874 A0A2P8XLK3 A0A182QD08 E0VHG5 A0A2M4BCT8 A0A2M4BCA0 A0A084VXD6 Q7PSQ5 V5GS79 W5JN50 A0A2M4AGP6 A0A182FLK3 A0A2M3Z242 A0A2M4AGR0 A0A2M3Z263 A0A146KTB8 A0A1I8N7F8 A0A182J144 A0A210QVH4 B4M812 A0A3B0KSD9 A0A1Y1N5V3 A0A0M9A5B9 B4H4F9 Q29I09 A0A088A5S0 W8BQ10 A0A0M4EP13 A0A3R7PVS9 B4JKJ5 A0A0P5Y5F1 A0A0P4Z516 A0A0P5B899 A0A0P5V869 U3JNW4 A0A0P5Q8S3 A0A0N8CCQ3 A0A0N8C2A5 A0A164K7R0 A0A0P6G1F3 A0A0P6HHW7 A0A0P6G852 V9K8G2 A0A1I8PZ32 A0A0P6CUV9 A0A0N8DVT0 A0A0K8U5B9 A0A0P4Z553 A0A0P4YXQ4 A0A0K8V6V2 A0A0P5Y2F6 A0A0P5N884 C3YRU1 A0A0A0AV20 A0A091VBE1 A0A026WKA5 A0A151WLQ7 A0A0P5IVF2 A0A2I0U480
EC Number
3.1.-.-
Pubmed
EMBL
AGBW02003464
OWR55551.1
ODYU01001281
SOQ37284.1
JTDY01003687
KOB69147.1
+ More
KK853176 KDR10287.1 GANO01000229 JAB59642.1 NEVH01006828 PNF36371.1 UFQS01000065 UFQT01000065 SSW98927.1 SSX19313.1 CVRI01000004 CRK87620.1 GEZM01012032 GEZM01012009 JAV93255.1 GFDF01003913 JAV10171.1 GFDL01001380 JAV33665.1 GFDL01001370 JAV33675.1 KQ971361 EFA08175.1 CH477976 EAT34679.1 JXUM01047499 KQ561521 KXJ78303.1 PYGN01001777 PSN32852.1 AXCN02002127 DS235170 EEB12821.1 GGFJ01001507 MBW50648.1 GGFJ01001506 MBW50647.1 ATLV01017988 KE525205 KFB42630.1 AAAB01008816 EAA05192.5 GALX01001457 JAB67009.1 ADMH02000630 ETN65551.1 GGFK01006642 MBW39963.1 GGFM01001820 MBW22571.1 GGFK01006653 MBW39974.1 GGFM01001843 MBW22594.1 GDHC01018898 JAP99730.1 NEDP02001668 OWF52730.1 CH940653 EDW62288.1 OUUW01000025 SPP89629.1 GEZM01012027 GEZM01012021 JAV93273.1 KQ435732 KOX77550.1 CH479209 EDW32710.1 CH379064 EAL31597.3 GAMC01005473 JAC01083.1 CP012528 ALC49896.1 QCYY01000591 ROT84134.1 CH916370 EDW00098.1 GDIP01063696 JAM40019.1 GDIP01217872 JAJ05530.1 GDIP01188920 JAJ34482.1 GDIP01102979 JAM00736.1 AGTO01010573 GDIQ01124217 JAL27509.1 GDIQ01092635 JAL59091.1 GDIQ01121819 JAL29907.1 LRGB01003375 KZS03025.1 GDIQ01039905 JAN54832.1 GDIQ01042599 JAN52138.1 GDIQ01039904 JAN54833.1 JW861536 AFO94053.1 GDIQ01087027 JAN07710.1 GDIQ01087028 JAN07709.1 GDHF01030578 JAI21736.1 GDIP01222166 JAJ01236.1 GDIP01222165 JAJ01237.1 GDHF01017703 JAI34611.1 GDIP01065482 JAM38233.1 GDIQ01148468 JAL03258.1 GG666548 EEN56846.1 KL872975 KGL97368.1 KK733824 KFR00095.1 KK107167 QOIP01000010 EZA56408.1 RLU17601.1 KQ982959 KYQ48773.1 GDIQ01207278 JAK44447.1 KZ506192 PKU40894.1
KK853176 KDR10287.1 GANO01000229 JAB59642.1 NEVH01006828 PNF36371.1 UFQS01000065 UFQT01000065 SSW98927.1 SSX19313.1 CVRI01000004 CRK87620.1 GEZM01012032 GEZM01012009 JAV93255.1 GFDF01003913 JAV10171.1 GFDL01001380 JAV33665.1 GFDL01001370 JAV33675.1 KQ971361 EFA08175.1 CH477976 EAT34679.1 JXUM01047499 KQ561521 KXJ78303.1 PYGN01001777 PSN32852.1 AXCN02002127 DS235170 EEB12821.1 GGFJ01001507 MBW50648.1 GGFJ01001506 MBW50647.1 ATLV01017988 KE525205 KFB42630.1 AAAB01008816 EAA05192.5 GALX01001457 JAB67009.1 ADMH02000630 ETN65551.1 GGFK01006642 MBW39963.1 GGFM01001820 MBW22571.1 GGFK01006653 MBW39974.1 GGFM01001843 MBW22594.1 GDHC01018898 JAP99730.1 NEDP02001668 OWF52730.1 CH940653 EDW62288.1 OUUW01000025 SPP89629.1 GEZM01012027 GEZM01012021 JAV93273.1 KQ435732 KOX77550.1 CH479209 EDW32710.1 CH379064 EAL31597.3 GAMC01005473 JAC01083.1 CP012528 ALC49896.1 QCYY01000591 ROT84134.1 CH916370 EDW00098.1 GDIP01063696 JAM40019.1 GDIP01217872 JAJ05530.1 GDIP01188920 JAJ34482.1 GDIP01102979 JAM00736.1 AGTO01010573 GDIQ01124217 JAL27509.1 GDIQ01092635 JAL59091.1 GDIQ01121819 JAL29907.1 LRGB01003375 KZS03025.1 GDIQ01039905 JAN54832.1 GDIQ01042599 JAN52138.1 GDIQ01039904 JAN54833.1 JW861536 AFO94053.1 GDIQ01087027 JAN07710.1 GDIQ01087028 JAN07709.1 GDHF01030578 JAI21736.1 GDIP01222166 JAJ01236.1 GDIP01222165 JAJ01237.1 GDHF01017703 JAI34611.1 GDIP01065482 JAM38233.1 GDIQ01148468 JAL03258.1 GG666548 EEN56846.1 KL872975 KGL97368.1 KK733824 KFR00095.1 KK107167 QOIP01000010 EZA56408.1 RLU17601.1 KQ982959 KYQ48773.1 GDIQ01207278 JAK44447.1 KZ506192 PKU40894.1
Proteomes
UP000007151
UP000037510
UP000027135
UP000192223
UP000235965
UP000183832
+ More
UP000007266 UP000008820 UP000069940 UP000249989 UP000245037 UP000075886 UP000009046 UP000030765 UP000007062 UP000000673 UP000069272 UP000095301 UP000075880 UP000242188 UP000008792 UP000268350 UP000053105 UP000008744 UP000001819 UP000005203 UP000092553 UP000283509 UP000001070 UP000016665 UP000076858 UP000095300 UP000001554 UP000053858 UP000053605 UP000053097 UP000279307 UP000075809
UP000007266 UP000008820 UP000069940 UP000249989 UP000245037 UP000075886 UP000009046 UP000030765 UP000007062 UP000000673 UP000069272 UP000095301 UP000075880 UP000242188 UP000008792 UP000268350 UP000053105 UP000008744 UP000001819 UP000005203 UP000092553 UP000283509 UP000001070 UP000016665 UP000076858 UP000095300 UP000001554 UP000053858 UP000053605 UP000053097 UP000279307 UP000075809
Pfam
PF07819 PGAP1
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
A0A212FP83
A0A2H1VAP4
A0A0L7L106
A0A067QZ37
A0A1W4WDY2
U5EZS7
+ More
A0A2J7R6B4 A0A336LRH5 A0A1J1HJN1 A0A1Y1N9E7 A0A1L8DUQ7 A0A1Q3G1I1 A0A1Q3G1J6 D6WW73 A0A1S4FY25 Q16K53 A0A182G874 A0A2P8XLK3 A0A182QD08 E0VHG5 A0A2M4BCT8 A0A2M4BCA0 A0A084VXD6 Q7PSQ5 V5GS79 W5JN50 A0A2M4AGP6 A0A182FLK3 A0A2M3Z242 A0A2M4AGR0 A0A2M3Z263 A0A146KTB8 A0A1I8N7F8 A0A182J144 A0A210QVH4 B4M812 A0A3B0KSD9 A0A1Y1N5V3 A0A0M9A5B9 B4H4F9 Q29I09 A0A088A5S0 W8BQ10 A0A0M4EP13 A0A3R7PVS9 B4JKJ5 A0A0P5Y5F1 A0A0P4Z516 A0A0P5B899 A0A0P5V869 U3JNW4 A0A0P5Q8S3 A0A0N8CCQ3 A0A0N8C2A5 A0A164K7R0 A0A0P6G1F3 A0A0P6HHW7 A0A0P6G852 V9K8G2 A0A1I8PZ32 A0A0P6CUV9 A0A0N8DVT0 A0A0K8U5B9 A0A0P4Z553 A0A0P4YXQ4 A0A0K8V6V2 A0A0P5Y2F6 A0A0P5N884 C3YRU1 A0A0A0AV20 A0A091VBE1 A0A026WKA5 A0A151WLQ7 A0A0P5IVF2 A0A2I0U480
A0A2J7R6B4 A0A336LRH5 A0A1J1HJN1 A0A1Y1N9E7 A0A1L8DUQ7 A0A1Q3G1I1 A0A1Q3G1J6 D6WW73 A0A1S4FY25 Q16K53 A0A182G874 A0A2P8XLK3 A0A182QD08 E0VHG5 A0A2M4BCT8 A0A2M4BCA0 A0A084VXD6 Q7PSQ5 V5GS79 W5JN50 A0A2M4AGP6 A0A182FLK3 A0A2M3Z242 A0A2M4AGR0 A0A2M3Z263 A0A146KTB8 A0A1I8N7F8 A0A182J144 A0A210QVH4 B4M812 A0A3B0KSD9 A0A1Y1N5V3 A0A0M9A5B9 B4H4F9 Q29I09 A0A088A5S0 W8BQ10 A0A0M4EP13 A0A3R7PVS9 B4JKJ5 A0A0P5Y5F1 A0A0P4Z516 A0A0P5B899 A0A0P5V869 U3JNW4 A0A0P5Q8S3 A0A0N8CCQ3 A0A0N8C2A5 A0A164K7R0 A0A0P6G1F3 A0A0P6HHW7 A0A0P6G852 V9K8G2 A0A1I8PZ32 A0A0P6CUV9 A0A0N8DVT0 A0A0K8U5B9 A0A0P4Z553 A0A0P4YXQ4 A0A0K8V6V2 A0A0P5Y2F6 A0A0P5N884 C3YRU1 A0A0A0AV20 A0A091VBE1 A0A026WKA5 A0A151WLQ7 A0A0P5IVF2 A0A2I0U480
Ontologies
PATHWAY
GO
PANTHER
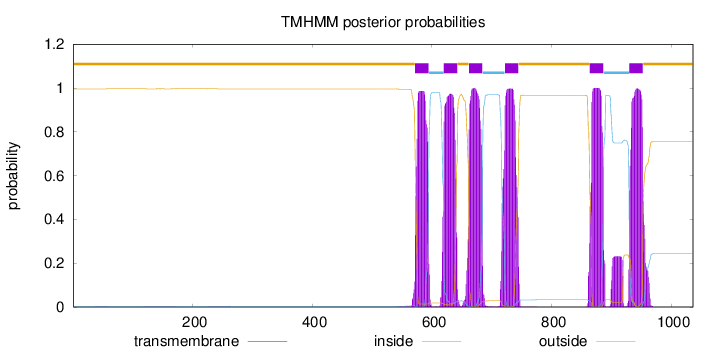
Topology
Subcellular location
Endoplasmic reticulum membrane
Length:
1036
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
139.82153
Exp number, first 60 AAs:
0.00499
Total prob of N-in:
0.00383
outside
1 - 571
TMhelix
572 - 594
inside
595 - 619
TMhelix
620 - 642
outside
643 - 661
TMhelix
662 - 684
inside
685 - 721
TMhelix
722 - 744
outside
745 - 863
TMhelix
864 - 886
inside
887 - 929
TMhelix
930 - 952
outside
953 - 1036
Population Genetic Test Statistics
Pi
256.121717
Theta
21.229776
Tajima's D
1.317998
CLR
0.320071
CSRT
0.749012549372531
Interpretation
Uncertain
