Gene
KWMTBOMO10376 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007061
Annotation
cathepsin_B_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.585 Lysosomal Reliability : 2.234
Sequence
CDS
ATGTTTATTTCGCGTGCGGCGTATGTTACGCTGGTGTGCGTTCTTGCAGCGGCCAAAGACTTACCGCATCCACTCTCCGATGAATTCATCAACACGATTAACCTAAAGCAAAATAGTTGGAAGGCGGGACGTAATTTTCCGCGCGACACATCGTTCGCGCATCTTAAGAAAATAATGGGAGTTATCGAGGATGAACATTTTGCGACCCTGCCAATAAAGACTCATAAAATCGATTTAATCGCCAGTCTGCCGGAAAACTTCGATCCCAGAGACAAATGGCCTGACTGTCCAACGTTGAATGAAGTCAGAGATCAAGGGTCTTGTGGCAGTTGTTGGGCTTTCGGTGCCGTCGAAGCTATGACAGACAGAGTATGTACCTATTCTAACGGAACTAAACATTTTCATTTTTCTGCCGAGGATTTGCTTAGTTGCTGCCCTATTTGTGGACTGGGATGCAGCGGAGGAATGCCGAGACTAGCTTGGGAATATTGGAAGCACTTCGGTCTAGTATCAGGAGGTAGTTACAATTCCAGTCAAGGTTGCAGACCTTACGAGATCCCTCCGTGTGAACATCACGTACCCGGCAACAGGATGCCCTGTAGCGGTGACACGAAGACTCCAAAATGCACAAAAAAATGCGAATCTGGATACGACGTTAATTACAAACAAGACAAACAATACGGAAAACATGTATATACTGTGTCCGGAGACGAAGACCACATCCGCGCGGAATTGTTCAAGAATGGTCCCGTCGAAGGTGCTTTCACAGTATATTCAGATTTGCTGTCGTACAAGAGTGGTGTATACAAACACACACAAGGGGACGCTCTCGGCGGGCACGCCGTCAAGATATTAGGCTGGGGTGTCGAAAATGATAACAAATATTGGCTTATTGCAAACTCGTGGAACTCAGATTGGGGAGACAATGGCTTCTTCAAAATTTTGCGCGGCGAGGATCACTGCGGTATTGAGAGCTCCATTGTTACGGGTGAACCGTTCTTAGATGAACACTAG
Protein
MFISRAAYVTLVCVLAAAKDLPHPLSDEFINTINLKQNSWKAGRNFPRDTSFAHLKKIMGVIEDEHFATLPIKTHKIDLIASLPENFDPRDKWPDCPTLNEVRDQGSCGSCWAFGAVEAMTDRVCTYSNGTKHFHFSAEDLLSCCPICGLGCSGGMPRLAWEYWKHFGLVSGGSYNSSQGCRPYEIPPCEHHVPGNRMPCSGDTKTPKCTKKCESGYDVNYKQDKQYGKHVYTVSGDEDHIRAELFKNGPVEGAFTVYSDLLSYKSGVYKHTQGDALGGHAVKILGWGVENDNKYWLIANSWNSDWGDNGFFKILRGEDHCGIESSIVTGEPFLDEH
Summary
Similarity
Belongs to the peptidase C1 family.
Uniprot
H9JC16
Q9BLI9
A1Z075
A0A194PJT4
A0A2A4J570
A0A0N1PH85
+ More
I4DIZ0 A1DYI5 A0A2H1WG36 I4DML4 E2J8A7 C7BWY6 Q5MGE8 Q9NHF5 A0A1E1VXU2 A0A0L7KTX3 S4NSX4 A0A212FP81 B2KSD9 B2KSE0 A0A2A4J768 A0A1V1FNM0 A0A0L7LR24 A0A1B0CXA8 A0A0T6B0A3 A0A336KB83 A0A1L8DPK1 A0A2A4J6I6 A0A1B0F0N2 B0W0V3 D6WGZ1 A0A0B5J1X8 A0A1J1IT42 U5EX33 A0A2H1W408 A0A2M3YZ69 A0A182SM11 Q70EX1 A0A1W4WLT1 A0A067RI52 A0A2P8XZP0 A0A1B6C2V6 A0A2M3YZ91 A0A1B6FMB6 W5JAL5 A0A182Y8P5 A0A182RE09 A0A2M4CFZ3 A0A2M4A1F2 A0A1B6CNK2 A0A1B6FGW4 A0A1B6G399 A0A1S4FMW6 A0A2J7Q2D7 A0A182QDS9 A0A2M3YZA5 A0A023EQ55 B4M3R5 Q1EGF0 A0A2J7Q2C6 A0A2M3YWH4 A0A2M4CGI0 A0A023EQ36 A0A1Y1LLE3 A0A084WTW2 A0A2M4A1I2 T1PA26 A0A182N9W5 A0A182JR86 A0A1Q3FN80 A0A2M4AIF0 A0A2M4A8M1 A0A2M4AGT9 T1DPY6 B4L388 A0A182F914 A0A0L7L6Z0 Q7Q9Y3 A0A182UK87 A0A182HYS5 A0A182W3J4 A0A2R7X282 A0A182XIF3 A0A2P2HVK2 A0A2H8TIP6 A0A182V7N7 A0A182JAJ1 A0A1I9WL58 A0A2M4BU45 A0A2M4BTS9 V9ISF8 A0A2K8JLG4 A0A2M4BT97 A0A2M4BTC9 A0A2M4BTM9 A0A1B6K5R8 A0A182M059 A9JSF8 A0A182GJJ7
I4DIZ0 A1DYI5 A0A2H1WG36 I4DML4 E2J8A7 C7BWY6 Q5MGE8 Q9NHF5 A0A1E1VXU2 A0A0L7KTX3 S4NSX4 A0A212FP81 B2KSD9 B2KSE0 A0A2A4J768 A0A1V1FNM0 A0A0L7LR24 A0A1B0CXA8 A0A0T6B0A3 A0A336KB83 A0A1L8DPK1 A0A2A4J6I6 A0A1B0F0N2 B0W0V3 D6WGZ1 A0A0B5J1X8 A0A1J1IT42 U5EX33 A0A2H1W408 A0A2M3YZ69 A0A182SM11 Q70EX1 A0A1W4WLT1 A0A067RI52 A0A2P8XZP0 A0A1B6C2V6 A0A2M3YZ91 A0A1B6FMB6 W5JAL5 A0A182Y8P5 A0A182RE09 A0A2M4CFZ3 A0A2M4A1F2 A0A1B6CNK2 A0A1B6FGW4 A0A1B6G399 A0A1S4FMW6 A0A2J7Q2D7 A0A182QDS9 A0A2M3YZA5 A0A023EQ55 B4M3R5 Q1EGF0 A0A2J7Q2C6 A0A2M3YWH4 A0A2M4CGI0 A0A023EQ36 A0A1Y1LLE3 A0A084WTW2 A0A2M4A1I2 T1PA26 A0A182N9W5 A0A182JR86 A0A1Q3FN80 A0A2M4AIF0 A0A2M4A8M1 A0A2M4AGT9 T1DPY6 B4L388 A0A182F914 A0A0L7L6Z0 Q7Q9Y3 A0A182UK87 A0A182HYS5 A0A182W3J4 A0A2R7X282 A0A182XIF3 A0A2P2HVK2 A0A2H8TIP6 A0A182V7N7 A0A182JAJ1 A0A1I9WL58 A0A2M4BU45 A0A2M4BTS9 V9ISF8 A0A2K8JLG4 A0A2M4BT97 A0A2M4BTC9 A0A2M4BTM9 A0A1B6K5R8 A0A182M059 A9JSF8 A0A182GJJ7
Pubmed
EMBL
BABH01014707
KM027254
AB045595
AIU94623.1
BAB40804.1
EF154237
+ More
ABM05925.1 KQ459603 KPI92989.1 NWSH01002924 PCG67287.1 KQ460328 KPJ15645.1 AK401258 BAM17880.1 EF068255 ABK90823.1 ODYU01008433 SOQ52039.1 AK402532 BAM19154.1 HQ110064 ADN19566.1 FM957999 CAX16634.1 AY829838 AAV91452.1 AF222788 AAF35867.2 GDQN01011563 JAT79491.1 JTDY01005708 KOB66713.1 GAIX01013867 JAA78693.1 AGBW02003464 OWR55554.1 EU126818 ACC66065.1 EU126819 ACC66066.1 PCG67290.1 FX985417 BAX07430.1 JTDY01000293 KOB77859.1 AJWK01033486 LJIG01022451 KRT80525.1 UFQS01000234 UFQT01000234 SSX01751.1 SSX22129.1 GFDF01005780 JAV08304.1 PCG67288.1 AJVK01019293 DS231819 EDS42770.1 KQ971321 EFA01289.1 KP303290 AJF94889.1 CVRI01000059 CRL03411.1 GANO01001145 JAB58726.1 ODYU01006189 SOQ47825.1 GGFM01000757 MBW21508.1 AJ583509 CAE47498.1 KK852667 KDR18937.1 PYGN01001112 PSN37482.1 GEDC01029441 JAS07857.1 GGFM01000824 MBW21575.1 GECZ01018424 JAS51345.1 ADMH02001667 ETN61492.1 GGFL01000084 MBW64262.1 GGFK01001260 MBW34581.1 GEDC01022407 JAS14891.1 GECZ01020323 JAS49446.1 GECZ01012865 JAS56904.1 NEVH01019371 PNF22739.1 AXCN02001202 GGFM01000858 MBW21609.1 GAPW01002413 JAC11185.1 CH940651 EDW65440.1 AY626233 CH477599 AAV31917.1 EAT38474.1 PNF22740.1 GGFF01000145 MBW20612.1 GGFL01000083 MBW64261.1 GAPW01002443 JAC11155.1 GEZM01054344 JAV73751.1 ATLV01026924 KE525420 KFB53656.1 GGFK01001177 MBW34498.1 KA645631 AFP60260.1 GFDL01006099 JAV28946.1 GGFK01007239 MBW40560.1 GGFK01003833 MBW37154.1 GGFK01006679 MBW40000.1 GAMD01001550 JAB00041.1 CH933810 EDW07016.1 JTDY01002528 KOB71237.1 AAAB01008898 EAA09183.5 APCN01005404 KK856497 PTY25904.1 IACF01000029 LAB65845.1 GFXV01002190 MBW13995.1 KU932242 APA33878.1 GGFJ01007137 MBW56278.1 GGFJ01007210 MBW56351.1 JQ780374 AFM82475.1 MF683380 ATU82521.1 GGFJ01007138 MBW56279.1 GGFJ01007205 MBW56346.1 GGFJ01007211 MBW56352.1 GECU01000917 JAT06790.1 AXCM01000198 ABLF02021036 ABLF02021045 ABLF02038382 ABLF02038385 ABLF02038399 BK006309 DAA06097.1 JXUM01001677 KQ560126 KXJ84332.1
ABM05925.1 KQ459603 KPI92989.1 NWSH01002924 PCG67287.1 KQ460328 KPJ15645.1 AK401258 BAM17880.1 EF068255 ABK90823.1 ODYU01008433 SOQ52039.1 AK402532 BAM19154.1 HQ110064 ADN19566.1 FM957999 CAX16634.1 AY829838 AAV91452.1 AF222788 AAF35867.2 GDQN01011563 JAT79491.1 JTDY01005708 KOB66713.1 GAIX01013867 JAA78693.1 AGBW02003464 OWR55554.1 EU126818 ACC66065.1 EU126819 ACC66066.1 PCG67290.1 FX985417 BAX07430.1 JTDY01000293 KOB77859.1 AJWK01033486 LJIG01022451 KRT80525.1 UFQS01000234 UFQT01000234 SSX01751.1 SSX22129.1 GFDF01005780 JAV08304.1 PCG67288.1 AJVK01019293 DS231819 EDS42770.1 KQ971321 EFA01289.1 KP303290 AJF94889.1 CVRI01000059 CRL03411.1 GANO01001145 JAB58726.1 ODYU01006189 SOQ47825.1 GGFM01000757 MBW21508.1 AJ583509 CAE47498.1 KK852667 KDR18937.1 PYGN01001112 PSN37482.1 GEDC01029441 JAS07857.1 GGFM01000824 MBW21575.1 GECZ01018424 JAS51345.1 ADMH02001667 ETN61492.1 GGFL01000084 MBW64262.1 GGFK01001260 MBW34581.1 GEDC01022407 JAS14891.1 GECZ01020323 JAS49446.1 GECZ01012865 JAS56904.1 NEVH01019371 PNF22739.1 AXCN02001202 GGFM01000858 MBW21609.1 GAPW01002413 JAC11185.1 CH940651 EDW65440.1 AY626233 CH477599 AAV31917.1 EAT38474.1 PNF22740.1 GGFF01000145 MBW20612.1 GGFL01000083 MBW64261.1 GAPW01002443 JAC11155.1 GEZM01054344 JAV73751.1 ATLV01026924 KE525420 KFB53656.1 GGFK01001177 MBW34498.1 KA645631 AFP60260.1 GFDL01006099 JAV28946.1 GGFK01007239 MBW40560.1 GGFK01003833 MBW37154.1 GGFK01006679 MBW40000.1 GAMD01001550 JAB00041.1 CH933810 EDW07016.1 JTDY01002528 KOB71237.1 AAAB01008898 EAA09183.5 APCN01005404 KK856497 PTY25904.1 IACF01000029 LAB65845.1 GFXV01002190 MBW13995.1 KU932242 APA33878.1 GGFJ01007137 MBW56278.1 GGFJ01007210 MBW56351.1 JQ780374 AFM82475.1 MF683380 ATU82521.1 GGFJ01007138 MBW56279.1 GGFJ01007205 MBW56346.1 GGFJ01007211 MBW56352.1 GECU01000917 JAT06790.1 AXCM01000198 ABLF02021036 ABLF02021045 ABLF02038382 ABLF02038385 ABLF02038399 BK006309 DAA06097.1 JXUM01001677 KQ560126 KXJ84332.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000037510
UP000007151
+ More
UP000092461 UP000092462 UP000002320 UP000007266 UP000183832 UP000075901 UP000192223 UP000027135 UP000245037 UP000000673 UP000076408 UP000075900 UP000235965 UP000075886 UP000008792 UP000008820 UP000030765 UP000095301 UP000075884 UP000075881 UP000009192 UP000069272 UP000007062 UP000075902 UP000075840 UP000075920 UP000076407 UP000075903 UP000075880 UP000075883 UP000007819 UP000069940 UP000249989
UP000092461 UP000092462 UP000002320 UP000007266 UP000183832 UP000075901 UP000192223 UP000027135 UP000245037 UP000000673 UP000076408 UP000075900 UP000235965 UP000075886 UP000008792 UP000008820 UP000030765 UP000095301 UP000075884 UP000075881 UP000009192 UP000069272 UP000007062 UP000075902 UP000075840 UP000075920 UP000076407 UP000075903 UP000075880 UP000075883 UP000007819 UP000069940 UP000249989
Interpro
ProteinModelPortal
H9JC16
Q9BLI9
A1Z075
A0A194PJT4
A0A2A4J570
A0A0N1PH85
+ More
I4DIZ0 A1DYI5 A0A2H1WG36 I4DML4 E2J8A7 C7BWY6 Q5MGE8 Q9NHF5 A0A1E1VXU2 A0A0L7KTX3 S4NSX4 A0A212FP81 B2KSD9 B2KSE0 A0A2A4J768 A0A1V1FNM0 A0A0L7LR24 A0A1B0CXA8 A0A0T6B0A3 A0A336KB83 A0A1L8DPK1 A0A2A4J6I6 A0A1B0F0N2 B0W0V3 D6WGZ1 A0A0B5J1X8 A0A1J1IT42 U5EX33 A0A2H1W408 A0A2M3YZ69 A0A182SM11 Q70EX1 A0A1W4WLT1 A0A067RI52 A0A2P8XZP0 A0A1B6C2V6 A0A2M3YZ91 A0A1B6FMB6 W5JAL5 A0A182Y8P5 A0A182RE09 A0A2M4CFZ3 A0A2M4A1F2 A0A1B6CNK2 A0A1B6FGW4 A0A1B6G399 A0A1S4FMW6 A0A2J7Q2D7 A0A182QDS9 A0A2M3YZA5 A0A023EQ55 B4M3R5 Q1EGF0 A0A2J7Q2C6 A0A2M3YWH4 A0A2M4CGI0 A0A023EQ36 A0A1Y1LLE3 A0A084WTW2 A0A2M4A1I2 T1PA26 A0A182N9W5 A0A182JR86 A0A1Q3FN80 A0A2M4AIF0 A0A2M4A8M1 A0A2M4AGT9 T1DPY6 B4L388 A0A182F914 A0A0L7L6Z0 Q7Q9Y3 A0A182UK87 A0A182HYS5 A0A182W3J4 A0A2R7X282 A0A182XIF3 A0A2P2HVK2 A0A2H8TIP6 A0A182V7N7 A0A182JAJ1 A0A1I9WL58 A0A2M4BU45 A0A2M4BTS9 V9ISF8 A0A2K8JLG4 A0A2M4BT97 A0A2M4BTC9 A0A2M4BTM9 A0A1B6K5R8 A0A182M059 A9JSF8 A0A182GJJ7
I4DIZ0 A1DYI5 A0A2H1WG36 I4DML4 E2J8A7 C7BWY6 Q5MGE8 Q9NHF5 A0A1E1VXU2 A0A0L7KTX3 S4NSX4 A0A212FP81 B2KSD9 B2KSE0 A0A2A4J768 A0A1V1FNM0 A0A0L7LR24 A0A1B0CXA8 A0A0T6B0A3 A0A336KB83 A0A1L8DPK1 A0A2A4J6I6 A0A1B0F0N2 B0W0V3 D6WGZ1 A0A0B5J1X8 A0A1J1IT42 U5EX33 A0A2H1W408 A0A2M3YZ69 A0A182SM11 Q70EX1 A0A1W4WLT1 A0A067RI52 A0A2P8XZP0 A0A1B6C2V6 A0A2M3YZ91 A0A1B6FMB6 W5JAL5 A0A182Y8P5 A0A182RE09 A0A2M4CFZ3 A0A2M4A1F2 A0A1B6CNK2 A0A1B6FGW4 A0A1B6G399 A0A1S4FMW6 A0A2J7Q2D7 A0A182QDS9 A0A2M3YZA5 A0A023EQ55 B4M3R5 Q1EGF0 A0A2J7Q2C6 A0A2M3YWH4 A0A2M4CGI0 A0A023EQ36 A0A1Y1LLE3 A0A084WTW2 A0A2M4A1I2 T1PA26 A0A182N9W5 A0A182JR86 A0A1Q3FN80 A0A2M4AIF0 A0A2M4A8M1 A0A2M4AGT9 T1DPY6 B4L388 A0A182F914 A0A0L7L6Z0 Q7Q9Y3 A0A182UK87 A0A182HYS5 A0A182W3J4 A0A2R7X282 A0A182XIF3 A0A2P2HVK2 A0A2H8TIP6 A0A182V7N7 A0A182JAJ1 A0A1I9WL58 A0A2M4BU45 A0A2M4BTS9 V9ISF8 A0A2K8JLG4 A0A2M4BT97 A0A2M4BTC9 A0A2M4BTM9 A0A1B6K5R8 A0A182M059 A9JSF8 A0A182GJJ7
PDB
1MIR
E-value=8.35977e-110,
Score=1014
Ontologies
PATHWAY
GO
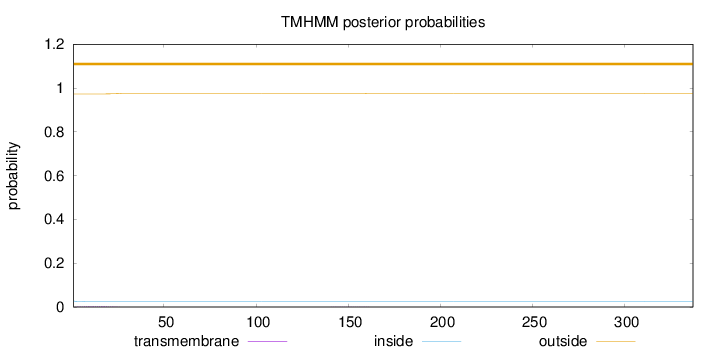
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.862393
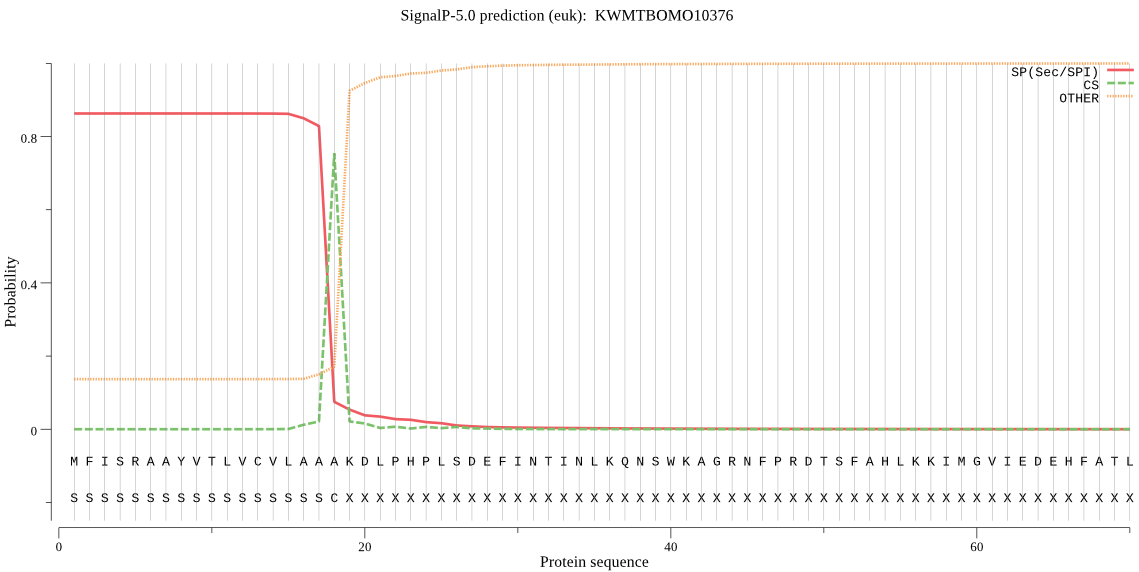
Length:
337
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06424
Exp number, first 60 AAs:
0.05739
Total prob of N-in:
0.02718
outside
1 - 337
Population Genetic Test Statistics
Pi
294.864459
Theta
206.735075
Tajima's D
1.333716
CLR
0
CSRT
0.747312634368282
Interpretation
Uncertain
