Gene
KWMTBOMO10374
Pre Gene Modal
BGIBMGA007046
Annotation
PREDICTED:_serine_protease_easter-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.075
Sequence
CDS
ATGACCCCACATGAACAACAGGGTTGGGTCACTAAATATCCTTGTTCAAGCCCTCATAATCATGGGAACAGCAGAGTTGGTGGTTTTACTGCGAATGCTAATGAAGGCTATATATGCTGCCCAAATGGTATTTGGGATCTAGAGAAACAAAACTCAGCCAATAATACTAAAGTGAATCCGGCCAGTGGTAATTTAGATCCGTTAATATCAAATCCTAAGCCGTCTCACCCCGAAGACCTTAGTGTTGACCAAGGAAAACCTAAACCACGAGTAAACGTTCTTATCCCGAGAACACCGCAGCCGACACTTGATCCTGGAACAAAAACCTCGTATTTTCCGAGTTTAAATAATAGCCCGGGAAATATAAACACATATAGCCAGTGCTCCCCATTAACAAGCTTTCCGCCTGATCCTAATACGGGGTGTTGTGGCCTGGAGGCTACTGAAACTGACAGGCTAACAGCCATCCCGAATACAATTTGGGCTCAGAAACCGTACATCCAAAATACTTGGAAACCTTTAAAACCCATGCTTGACCCATCAATACCATTATGGCAGAATAGTCAGAGTCATATCAATCGGAAGAAAAGAGAATTAATTAAGTCGTCGACTCGCATCGCAGGTGGTACCGAAACAGAGTTATATCAATATCCGTGGACAGTTCTGTTGAAAACGACATTCAATTATGGGACCCACAAAGCGGCGTTCAATTGTGGAGGATCTCTCATCAGTTCTCGCTACGTACTCACCGCCGGACATTGTGTGTTTGATCCAAACGGCACAATAGCCAATGTTGAAGTTCATCTAGCAGAATACGATAAGCAATCGTTTCCGGTCGACTGCAAAATGGTCTTAGGTGTCGGTCAGAAGTGCATACAAAATTTAATAATGTACGGAGAAGATGTTATTCGACACCCCCTATATGACGACAATAGCCTCAGTAATGACATCGCATTAATCAGACTACGTGGGAACGTTCCATATACAGATTATATTCGACCGATTTGTCTGCCCCCGATCAATATTGACGATCCGGAGTTTTCGAATTTGCGATTAGCGGTAGCTGGATGGGGTAGTAACGGTCGGTTTATATCTGACATCAAACAATCGACGGTCGTCAATCTAGTGCCACAGGCGCGATGCAAATTTTTTTATCCGGCTCTGAGCGACACACAGCTCTGTGCCGCCGGGTATTCTGGAGAGGACACATGCAAAGGCGACTCCGGAGGACCCTTAATGACCGTCTACGGCGGGAAGTACTTTGTCGTGGGTGTAGTGAGTGGAAAACGGGCTGACTCCCCCTGTGGGACATCCATGCCTTCGCTTTATAGTAACGTCTACCGGTACATAGACTGGATTCGGAGTAACATTCGTAATTAA
Protein
MTPHEQQGWVTKYPCSSPHNHGNSRVGGFTANANEGYICCPNGIWDLEKQNSANNTKVNPASGNLDPLISNPKPSHPEDLSVDQGKPKPRVNVLIPRTPQPTLDPGTKTSYFPSLNNSPGNINTYSQCSPLTSFPPDPNTGCCGLEATETDRLTAIPNTIWAQKPYIQNTWKPLKPMLDPSIPLWQNSQSHINRKKRELIKSSTRIAGGTETELYQYPWTVLLKTTFNYGTHKAAFNCGGSLISSRYVLTAGHCVFDPNGTIANVEVHLAEYDKQSFPVDCKMVLGVGQKCIQNLIMYGEDVIRHPLYDDNSLSNDIALIRLRGNVPYTDYIRPICLPPINIDDPEFSNLRLAVAGWGSNGRFISDIKQSTVVNLVPQARCKFFYPALSDTQLCAAGYSGEDTCKGDSGGPLMTVYGGKYFVVGVVSGKRADSPCGTSMPSLYSNVYRYIDWIRSNIRN
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
EMBL
BABH01014680
NWSH01001431
PCG71243.1
ODYU01003135
SOQ41515.1
RSAL01000036
+ More
RVE51294.1 AGBW02013343 OWR43557.1 KQ459603 KPI92987.1 JTDY01001765 KOB72956.1 KQ461155 KPJ08364.1 KY680241 ATD13322.1 JTDY01003981 KOB68749.1 NWSH01000467 PCG76208.1 ODYU01011137 SOQ56686.1 AY677082 AAW24481.1 AY077643 AAL76085.1 DQ115323 AAZ91696.1
RVE51294.1 AGBW02013343 OWR43557.1 KQ459603 KPI92987.1 JTDY01001765 KOB72956.1 KQ461155 KPJ08364.1 KY680241 ATD13322.1 JTDY01003981 KOB68749.1 NWSH01000467 PCG76208.1 ODYU01011137 SOQ56686.1 AY677082 AAW24481.1 AY077643 AAL76085.1 DQ115323 AAZ91696.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF50494
SSF50494
Gene 3D
CDD
ProteinModelPortal
PDB
2OLG
E-value=1.83957e-30,
Score=331
Ontologies
GO
Topology
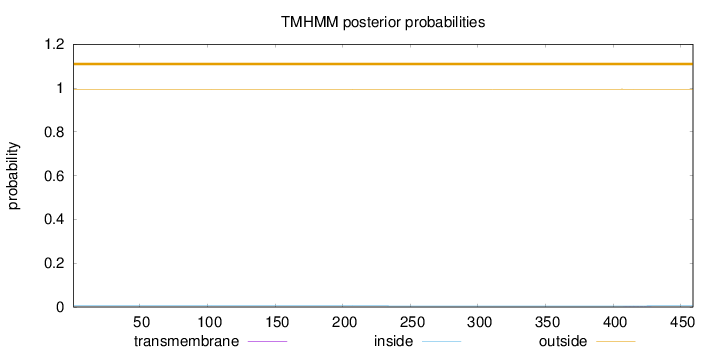
Length:
459
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01307
Exp number, first 60 AAs:
0.0001
Total prob of N-in:
0.00516
outside
1 - 459
Population Genetic Test Statistics
Pi
226.09612
Theta
206.232463
Tajima's D
0.383783
CLR
0
CSRT
0.478926053697315
Interpretation
Uncertain
