Gene
KWMTBOMO10373
Pre Gene Modal
BGIBMGA007047
Annotation
short-chain_dehydrogenase/reductase_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 3.053
Sequence
CDS
ATGTCCGTTAGCCGTCTTGTAGCTGTAACAGGGTGTGATAGTGGACTCGGTTGGGCAGTAGCAGCTCGATTAGCCCGAGAAGGCTTCATCACAATAGCTGGTATGCACAAAGGAATAGAAACCGAAGCAGCGAAAGCCCTCGAAAAATTGTGTGCGCACACATTCCCTTTAGACGTAACTAGAGTTGAAAGCGTTCAAGAATTCCGGAAGTACGTGCTTACAGTCTTGAAAGATAATCCTAAATATAAATTTCATGCCGTGGTCAACAACGCTGGAGTTATGACAGTAGGAAAGTACGAATGGATGACAGCATCGATGATAGAAAGCCCGGTTCATGTGAATTTGCTCGGCGCGATGAGAGTCATTTCCGCATTTTTACCAGAGATCAGAAAAACTGCAATCGAAACTACAAGCAAACCTAAGCCCCGTATAATCAACGTTGGTAGCCATTGTGGCCTGCAGCCTTTGCCCGCCTTCGCTGCATACAGTGCAAGCAAAGCTGGCCTTTTGGCGTTGACACGATGTCTACATCTTGAGCACAGCGAGCACGGATTAGCTGTCATTGCCTTTGTACCAGGTGGCTTCGTTGGCTCTAGTAACATTCTGCTCGGGCAAGAAACAAAAGGAGAAGCAATGGTTGAACATTTAAACGATGAACAACGAAAGTTTTACGGAAATAAAATTGAATCTCTAAACAATTATTTGGAGCTGGCATCTGGCGAGGGGAAATTTGATTCAATGCATGATATGAAAATATTGAACACTTTTATGAAGGCTATGTTGGATGAAACTCCGAAATTAATGTATAAAGTGGAATCCTGGAGATATATGTTTTATTATAATTTAATGAGATTGCCTTTGCCATTATTCATTCATAAATGGATTATTAAACAGTTTCTAAGCTTCCCTGATCACCAATGA
Protein
MSVSRLVAVTGCDSGLGWAVAARLAREGFITIAGMHKGIETEAAKALEKLCAHTFPLDVTRVESVQEFRKYVLTVLKDNPKYKFHAVVNNAGVMTVGKYEWMTASMIESPVHVNLLGAMRVISAFLPEIRKTAIETTSKPKPRIINVGSHCGLQPLPAFAAYSASKAGLLALTRCLHLEHSEHGLAVIAFVPGGFVGSSNILLGQETKGEAMVEHLNDEQRKFYGNKIESLNNYLELASGEGKFDSMHDMKILNTFMKAMLDETPKLMYKVESWRYMFYYNLMRLPLPLFIHKWIIKQFLSFPDHQ
Summary
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Uniprot
D4Q9I1
A0A2A4JI37
A0A060Q0K2
A0A194PHU2
A0A194QS64
A0A3S2M4M5
+ More
A0A212EPZ0 S4P318 K7J306 A0A1I8PCK7 A0A1I8N9B3 A0A182HAP1 E0VSJ3 A0A034WB66 A0A1I8N9A9 A0A0K8UPW8 A0A1Q3FS61 A0A1Y1LKZ5 A0A336M4K6 A0A1J1I8B0 B3MTJ0 A0A087ZTA4 A0A1B0A8W3 A0A1B0BZ77 A0A232EMD8 U4TY41 E9I9H5 J3JYH1 A0A1A9W6I3 A0A026W555 A0A195BX87 D6WSF7 B4R091 B4HZE9 A0A151IUF5 I3VPX6 Q9VAH3 A0A336LC86 B4PPK1 A0A2P8YI12 B4NBL7 B3P5D1 B0WWJ3 A0A0M9A4I9 A0A154P594 A0A151WUN3 A0A195CBN8 A0A067RSA4 A0A1W4URT6 A0A1A9UQ63 A0A1A9XMW0 A0A0M4EL31 Q29AP2 A0A1B6DNT4 B4G651 A0A1B6FUE5 B4M5T8 A0A2J7R9Z7 B4JYN4 B4K2K6 A0A1I8JUU4 A0A158NXK4 A0A0L7QM99 A0A182JGN0 B4K618 A0A1B6J899 A0A182QQW8 A0A182TRQ8 E2AIZ7 A0A182V2E1 A0A3B0K4J8 T1I0L0 A0A182WY31 Q7Q4T3 A0A146KVZ1 A0A0A9YUM4 A0A182HMT2 A0A182N0H3 A0A182K110 A0A182VV49 A0A2R7WFZ3 A0A034WB62 A0A0J7KS76 W5JT06 A0A182Y4T8 A0A182FIE9 J9KX18 A0A182KWA1 A0A182PL04 A0A0P5G1M5 A0A0P5J2U1 A0A0P5FYV4 A0A0P5RF26
A0A212EPZ0 S4P318 K7J306 A0A1I8PCK7 A0A1I8N9B3 A0A182HAP1 E0VSJ3 A0A034WB66 A0A1I8N9A9 A0A0K8UPW8 A0A1Q3FS61 A0A1Y1LKZ5 A0A336M4K6 A0A1J1I8B0 B3MTJ0 A0A087ZTA4 A0A1B0A8W3 A0A1B0BZ77 A0A232EMD8 U4TY41 E9I9H5 J3JYH1 A0A1A9W6I3 A0A026W555 A0A195BX87 D6WSF7 B4R091 B4HZE9 A0A151IUF5 I3VPX6 Q9VAH3 A0A336LC86 B4PPK1 A0A2P8YI12 B4NBL7 B3P5D1 B0WWJ3 A0A0M9A4I9 A0A154P594 A0A151WUN3 A0A195CBN8 A0A067RSA4 A0A1W4URT6 A0A1A9UQ63 A0A1A9XMW0 A0A0M4EL31 Q29AP2 A0A1B6DNT4 B4G651 A0A1B6FUE5 B4M5T8 A0A2J7R9Z7 B4JYN4 B4K2K6 A0A1I8JUU4 A0A158NXK4 A0A0L7QM99 A0A182JGN0 B4K618 A0A1B6J899 A0A182QQW8 A0A182TRQ8 E2AIZ7 A0A182V2E1 A0A3B0K4J8 T1I0L0 A0A182WY31 Q7Q4T3 A0A146KVZ1 A0A0A9YUM4 A0A182HMT2 A0A182N0H3 A0A182K110 A0A182VV49 A0A2R7WFZ3 A0A034WB62 A0A0J7KS76 W5JT06 A0A182Y4T8 A0A182FIE9 J9KX18 A0A182KWA1 A0A182PL04 A0A0P5G1M5 A0A0P5J2U1 A0A0P5FYV4 A0A0P5RF26
Pubmed
19121390
20501590
26354079
22118469
23622113
20075255
+ More
25315136 26483478 20566863 25348373 28004739 17994087 28648823 23537049 21282665 22516182 24508170 30249741 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 29403074 24845553 15632085 21347285 20798317 12364791 14747013 17210077 26823975 25401762 20920257 23761445 25244985 20966253
25315136 26483478 20566863 25348373 28004739 17994087 28648823 23537049 21282665 22516182 24508170 30249741 18362917 19820115 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 29403074 24845553 15632085 21347285 20798317 12364791 14747013 17210077 26823975 25401762 20920257 23761445 25244985 20966253
EMBL
BABH01014679
BABH01014680
AB361434
BAJ05088.1
NWSH01001431
PCG71244.1
+ More
AB894824 BAP01458.1 KQ459603 KPI92986.1 KQ461155 KPJ08363.1 RSAL01000036 RVE51295.1 AGBW02013343 OWR43558.1 GAIX01006669 JAA85891.1 JXUM01123026 KQ566670 KXJ69924.1 DS235751 EEB16349.1 GAKP01007008 GAKP01007005 GAKP01007003 JAC51944.1 GDHF01023600 JAI28714.1 GFDL01004638 JAV30407.1 GEZM01052703 JAV74313.1 UFQS01000550 UFQT01000550 SSX04833.1 SSX25196.1 CVRI01000039 CRK94649.1 CH902623 EDV30580.1 JXJN01023016 NNAY01003399 OXU19472.1 KB631818 ERL86534.1 GL761811 EFZ22778.1 BT128299 AEE63257.1 KK107405 QOIP01000014 EZA51210.1 RLU14939.1 KQ976394 KYM93254.1 KQ971352 EFA06375.2 CM000364 EDX14897.1 CH480819 EDW53406.1 KQ980960 KYN11053.1 BT149840 AFK83748.1 AE014297 AB361435 AAF56937.1 BAJ05089.1 UFQS01002626 UFQT01002626 SSX14387.1 SSX33798.1 CM000160 EDW98251.1 PYGN01000580 PSN43891.1 CH964232 EDW81181.1 CH954182 EDV53181.1 DS232148 EDS36085.1 KQ435736 KOX77077.1 KQ434809 KZC06504.1 KQ982730 KYQ51538.1 KQ978068 KYM97616.1 KK852498 KDR22689.1 CP012526 ALC48046.1 CM000070 EAL27307.2 GEDC01009978 GEDC01000525 JAS27320.1 JAS36773.1 CH479179 EDW23810.1 GECZ01015964 JAS53805.1 CH940652 EDW59014.1 NEVH01006580 PNF37650.1 CH916377 EDV90796.1 CH919373 EDW04586.1 ADTU01030685 KQ414894 KOC59747.1 CH933806 EDW16255.1 GECU01012308 JAS95398.1 AXCN02001411 GL439921 EFN66586.1 OUUW01000007 SPP82920.1 ACPB03008738 AAAB01008963 EAA12045.4 GDHC01017965 JAQ00664.1 GBHO01006907 GBHO01006906 GBHO01006904 JAG36697.1 JAG36698.1 JAG36700.1 APCN01004903 KK854724 PTY18191.1 GAKP01007013 JAC51939.1 LBMM01003833 KMQ93074.1 ADMH02000457 ETN66403.1 ABLF02029076 GDIQ01247656 JAK04069.1 GDIQ01206268 JAK45457.1 GDIQ01249309 JAK02416.1 GDIQ01101654 JAL50072.1
AB894824 BAP01458.1 KQ459603 KPI92986.1 KQ461155 KPJ08363.1 RSAL01000036 RVE51295.1 AGBW02013343 OWR43558.1 GAIX01006669 JAA85891.1 JXUM01123026 KQ566670 KXJ69924.1 DS235751 EEB16349.1 GAKP01007008 GAKP01007005 GAKP01007003 JAC51944.1 GDHF01023600 JAI28714.1 GFDL01004638 JAV30407.1 GEZM01052703 JAV74313.1 UFQS01000550 UFQT01000550 SSX04833.1 SSX25196.1 CVRI01000039 CRK94649.1 CH902623 EDV30580.1 JXJN01023016 NNAY01003399 OXU19472.1 KB631818 ERL86534.1 GL761811 EFZ22778.1 BT128299 AEE63257.1 KK107405 QOIP01000014 EZA51210.1 RLU14939.1 KQ976394 KYM93254.1 KQ971352 EFA06375.2 CM000364 EDX14897.1 CH480819 EDW53406.1 KQ980960 KYN11053.1 BT149840 AFK83748.1 AE014297 AB361435 AAF56937.1 BAJ05089.1 UFQS01002626 UFQT01002626 SSX14387.1 SSX33798.1 CM000160 EDW98251.1 PYGN01000580 PSN43891.1 CH964232 EDW81181.1 CH954182 EDV53181.1 DS232148 EDS36085.1 KQ435736 KOX77077.1 KQ434809 KZC06504.1 KQ982730 KYQ51538.1 KQ978068 KYM97616.1 KK852498 KDR22689.1 CP012526 ALC48046.1 CM000070 EAL27307.2 GEDC01009978 GEDC01000525 JAS27320.1 JAS36773.1 CH479179 EDW23810.1 GECZ01015964 JAS53805.1 CH940652 EDW59014.1 NEVH01006580 PNF37650.1 CH916377 EDV90796.1 CH919373 EDW04586.1 ADTU01030685 KQ414894 KOC59747.1 CH933806 EDW16255.1 GECU01012308 JAS95398.1 AXCN02001411 GL439921 EFN66586.1 OUUW01000007 SPP82920.1 ACPB03008738 AAAB01008963 EAA12045.4 GDHC01017965 JAQ00664.1 GBHO01006907 GBHO01006906 GBHO01006904 JAG36697.1 JAG36698.1 JAG36700.1 APCN01004903 KK854724 PTY18191.1 GAKP01007013 JAC51939.1 LBMM01003833 KMQ93074.1 ADMH02000457 ETN66403.1 ABLF02029076 GDIQ01247656 JAK04069.1 GDIQ01206268 JAK45457.1 GDIQ01249309 JAK02416.1 GDIQ01101654 JAL50072.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000283053
UP000007151
+ More
UP000002358 UP000095300 UP000095301 UP000069940 UP000249989 UP000009046 UP000183832 UP000007801 UP000005203 UP000092445 UP000092460 UP000215335 UP000030742 UP000091820 UP000053097 UP000279307 UP000078540 UP000007266 UP000000304 UP000001292 UP000078492 UP000000803 UP000002282 UP000245037 UP000007798 UP000008711 UP000002320 UP000053105 UP000076502 UP000075809 UP000078542 UP000027135 UP000192221 UP000078200 UP000092443 UP000092553 UP000001819 UP000008744 UP000008792 UP000235965 UP000001070 UP000075900 UP000005205 UP000053825 UP000075880 UP000009192 UP000075886 UP000075902 UP000000311 UP000075903 UP000268350 UP000015103 UP000076407 UP000007062 UP000075840 UP000075884 UP000075881 UP000075920 UP000036403 UP000000673 UP000076408 UP000069272 UP000007819 UP000075882 UP000075885
UP000002358 UP000095300 UP000095301 UP000069940 UP000249989 UP000009046 UP000183832 UP000007801 UP000005203 UP000092445 UP000092460 UP000215335 UP000030742 UP000091820 UP000053097 UP000279307 UP000078540 UP000007266 UP000000304 UP000001292 UP000078492 UP000000803 UP000002282 UP000245037 UP000007798 UP000008711 UP000002320 UP000053105 UP000076502 UP000075809 UP000078542 UP000027135 UP000192221 UP000078200 UP000092443 UP000092553 UP000001819 UP000008744 UP000008792 UP000235965 UP000001070 UP000075900 UP000005205 UP000053825 UP000075880 UP000009192 UP000075886 UP000075902 UP000000311 UP000075903 UP000268350 UP000015103 UP000076407 UP000007062 UP000075840 UP000075884 UP000075881 UP000075920 UP000036403 UP000000673 UP000076408 UP000069272 UP000007819 UP000075882 UP000075885
Pfam
PF00106 adh_short
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
D4Q9I1
A0A2A4JI37
A0A060Q0K2
A0A194PHU2
A0A194QS64
A0A3S2M4M5
+ More
A0A212EPZ0 S4P318 K7J306 A0A1I8PCK7 A0A1I8N9B3 A0A182HAP1 E0VSJ3 A0A034WB66 A0A1I8N9A9 A0A0K8UPW8 A0A1Q3FS61 A0A1Y1LKZ5 A0A336M4K6 A0A1J1I8B0 B3MTJ0 A0A087ZTA4 A0A1B0A8W3 A0A1B0BZ77 A0A232EMD8 U4TY41 E9I9H5 J3JYH1 A0A1A9W6I3 A0A026W555 A0A195BX87 D6WSF7 B4R091 B4HZE9 A0A151IUF5 I3VPX6 Q9VAH3 A0A336LC86 B4PPK1 A0A2P8YI12 B4NBL7 B3P5D1 B0WWJ3 A0A0M9A4I9 A0A154P594 A0A151WUN3 A0A195CBN8 A0A067RSA4 A0A1W4URT6 A0A1A9UQ63 A0A1A9XMW0 A0A0M4EL31 Q29AP2 A0A1B6DNT4 B4G651 A0A1B6FUE5 B4M5T8 A0A2J7R9Z7 B4JYN4 B4K2K6 A0A1I8JUU4 A0A158NXK4 A0A0L7QM99 A0A182JGN0 B4K618 A0A1B6J899 A0A182QQW8 A0A182TRQ8 E2AIZ7 A0A182V2E1 A0A3B0K4J8 T1I0L0 A0A182WY31 Q7Q4T3 A0A146KVZ1 A0A0A9YUM4 A0A182HMT2 A0A182N0H3 A0A182K110 A0A182VV49 A0A2R7WFZ3 A0A034WB62 A0A0J7KS76 W5JT06 A0A182Y4T8 A0A182FIE9 J9KX18 A0A182KWA1 A0A182PL04 A0A0P5G1M5 A0A0P5J2U1 A0A0P5FYV4 A0A0P5RF26
A0A212EPZ0 S4P318 K7J306 A0A1I8PCK7 A0A1I8N9B3 A0A182HAP1 E0VSJ3 A0A034WB66 A0A1I8N9A9 A0A0K8UPW8 A0A1Q3FS61 A0A1Y1LKZ5 A0A336M4K6 A0A1J1I8B0 B3MTJ0 A0A087ZTA4 A0A1B0A8W3 A0A1B0BZ77 A0A232EMD8 U4TY41 E9I9H5 J3JYH1 A0A1A9W6I3 A0A026W555 A0A195BX87 D6WSF7 B4R091 B4HZE9 A0A151IUF5 I3VPX6 Q9VAH3 A0A336LC86 B4PPK1 A0A2P8YI12 B4NBL7 B3P5D1 B0WWJ3 A0A0M9A4I9 A0A154P594 A0A151WUN3 A0A195CBN8 A0A067RSA4 A0A1W4URT6 A0A1A9UQ63 A0A1A9XMW0 A0A0M4EL31 Q29AP2 A0A1B6DNT4 B4G651 A0A1B6FUE5 B4M5T8 A0A2J7R9Z7 B4JYN4 B4K2K6 A0A1I8JUU4 A0A158NXK4 A0A0L7QM99 A0A182JGN0 B4K618 A0A1B6J899 A0A182QQW8 A0A182TRQ8 E2AIZ7 A0A182V2E1 A0A3B0K4J8 T1I0L0 A0A182WY31 Q7Q4T3 A0A146KVZ1 A0A0A9YUM4 A0A182HMT2 A0A182N0H3 A0A182K110 A0A182VV49 A0A2R7WFZ3 A0A034WB62 A0A0J7KS76 W5JT06 A0A182Y4T8 A0A182FIE9 J9KX18 A0A182KWA1 A0A182PL04 A0A0P5G1M5 A0A0P5J2U1 A0A0P5FYV4 A0A0P5RF26
PDB
3V2H
E-value=4.41082e-11,
Score=162
Ontologies
GO
Topology
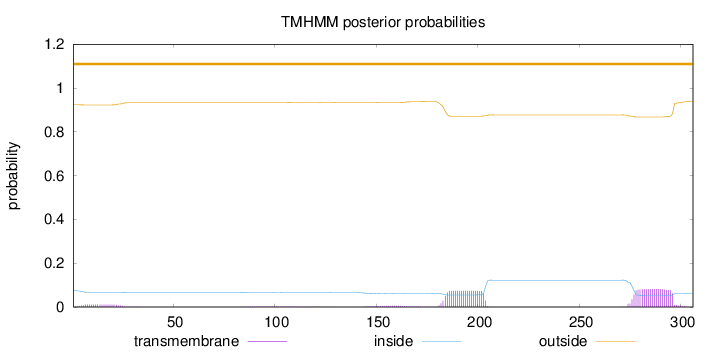
Length:
306
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.56768
Exp number, first 60 AAs:
0.22451
Total prob of N-in:
0.07620
outside
1 - 306
Population Genetic Test Statistics
Pi
24.470394
Theta
20.989635
Tajima's D
1.132332
CLR
0.021133
CSRT
0.702514874256287
Interpretation
Uncertain
