Gene
KWMTBOMO10371
Pre Gene Modal
BGIBMGA007050
Annotation
PREDICTED:_TM2_domain-containing_protein_CG10795_[Plutella_xylostella]
Full name
TM2 domain-containing protein CG10795
Location in the cell
PlasmaMembrane Reliability : 2.338
Sequence
CDS
ATGATGAAACTTGTACATTCTGTACACGTCTACGTATGTTTGGTATTTTTGCTATCGATTACAGAAGCTTCCGATGAAGCTAAAGTTGATAAAGTGCATGTAGACTGCAGTACGTTAAGATTGGGACAATTTATTTGTCCAAACCCAAGTATTGATCAAATAGATCCAGACACACAACAATTCAGAGGATGTATAAAAGGAAAGGAAATTCCGTCCGAAGGTGAAGCCGAAGTGGAGTGTATAGCAGCTGATGGAATAATATGTAATGAAACAAAAAATGGAACATTTACAAGGAAACTACCATGTAAATGGACGAATGGATATTCTCTAGAAATTGCTTTATTACTATCTGTATTCCTGGGAATGTTTGGATTGGATCGCTTTTATTTGGGATATCCTGGAATGGGTTTAGCAAAACTATGTACTCTGGGATTCATGTTCCTCGGACAGTTATTAGACATTATATTAATAGCTGCACAGGTTGTGGGACCTTCAGATGGGTCTGCTTATGTCATTCCATATTATGGTGCTGGAGTTACTGTTGTTCGGAGTGATAATGAGACTTACCTACTACCACAACCAGATTGGCATAACATCACCTGA
Protein
MMKLVHSVHVYVCLVFLLSITEASDEAKVDKVHVDCSTLRLGQFICPNPSIDQIDPDTQQFRGCIKGKEIPSEGEAEVECIAADGIICNETKNGTFTRKLPCKWTNGYSLEIALLLSVFLGMFGLDRFYLGYPGMGLAKLCTLGFMFLGQLLDIILIAAQVVGPSDGSAYVIPYYGAGVTVVRSDNETYLLPQPDWHNIT
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Belongs to the TM2 family.
Belongs to the TM2 family.
Keywords
Complete proteome
Glycoprotein
Membrane
Reference proteome
Signal
Transmembrane
Transmembrane helix
Feature
chain TM2 domain-containing protein CG10795
Uniprot
H9JC05
A0A2A4JHQ4
A0A2H1VQ05
S4PMA3
A0A194QXQ5
A0A194PJS8
+ More
A0A0L7LN27 A0A3S2P3B3 A0A212F5G3 A0A2M4C0I7 A0A182SAP2 A0A182MEI2 A0A084WNB0 A0A182S4P0 B0X5T7 A0A2M4CHQ6 A0A182JB19 D6WRM4 T1DQ81 A0A1Q3FJ65 A0A023EJ15 U5EWP4 Q16N92 A0A1S4FV38 A0A182KRB5 Q7QJF1 A0A2M4CML1 A0A067RDS3 A0A0K8U034 A0A034WVA9 A0A3B0J1K0 A0A0A1XS12 A0A1B0CIK9 Q28WL7 B4H6Z5 A0A182JUH7 A0A182WJ80 A0A182NHV2 A0A232FAA0 A0A182PQE9 W5JTA3 J3JWZ5 E0VQH1 A0A0A9YU71 A0A182FF46 A0A158NCG2 A0A182XYW6 A0A2P8YXF5 A0A1L8D9A6 A0A1L8D8X2 A0A182VHI4 A0A1W4VXS6 K7J455 B3NMV8 A0A182I3B1 A0A182X3Q8 B4I7P9 B4QG30 B4MJU8 Q9W2H1 A0A1B6K4I5 A0A0N7Z8M7 T1HXJ1 A0A1B6DVI9 B4P8Z6 B4LLH2 W8CA92 B3MBY1 A0A1W4W6Q6 A0A151JQW3 A0A151WMZ4 B4KRF3 A0A2S2QEV5 A0A0V0G9B3 A0A026X170 A0A224Y076 A0A087ZV56 B4J6S7 A0A0M5JAA8 A0A1S4E8X3 A0A3Q0ISJ9 A0A1I8MII9 A0A0L0C310 A0A170Y667 E2B678 N6TLB6 U4U6Q3 A0A195F9H6 A0A2H8TZC0 C4WUJ5 A0A1I8NNX6 A0A1A9WJX4 A0A0L7R1D7 E9INI5 A0A1A9V211 A0A1B0BY58 A0A2A3E631 F4WX05 A0A1A9XB89 E1ZWA9
A0A0L7LN27 A0A3S2P3B3 A0A212F5G3 A0A2M4C0I7 A0A182SAP2 A0A182MEI2 A0A084WNB0 A0A182S4P0 B0X5T7 A0A2M4CHQ6 A0A182JB19 D6WRM4 T1DQ81 A0A1Q3FJ65 A0A023EJ15 U5EWP4 Q16N92 A0A1S4FV38 A0A182KRB5 Q7QJF1 A0A2M4CML1 A0A067RDS3 A0A0K8U034 A0A034WVA9 A0A3B0J1K0 A0A0A1XS12 A0A1B0CIK9 Q28WL7 B4H6Z5 A0A182JUH7 A0A182WJ80 A0A182NHV2 A0A232FAA0 A0A182PQE9 W5JTA3 J3JWZ5 E0VQH1 A0A0A9YU71 A0A182FF46 A0A158NCG2 A0A182XYW6 A0A2P8YXF5 A0A1L8D9A6 A0A1L8D8X2 A0A182VHI4 A0A1W4VXS6 K7J455 B3NMV8 A0A182I3B1 A0A182X3Q8 B4I7P9 B4QG30 B4MJU8 Q9W2H1 A0A1B6K4I5 A0A0N7Z8M7 T1HXJ1 A0A1B6DVI9 B4P8Z6 B4LLH2 W8CA92 B3MBY1 A0A1W4W6Q6 A0A151JQW3 A0A151WMZ4 B4KRF3 A0A2S2QEV5 A0A0V0G9B3 A0A026X170 A0A224Y076 A0A087ZV56 B4J6S7 A0A0M5JAA8 A0A1S4E8X3 A0A3Q0ISJ9 A0A1I8MII9 A0A0L0C310 A0A170Y667 E2B678 N6TLB6 U4U6Q3 A0A195F9H6 A0A2H8TZC0 C4WUJ5 A0A1I8NNX6 A0A1A9WJX4 A0A0L7R1D7 E9INI5 A0A1A9V211 A0A1B0BY58 A0A2A3E631 F4WX05 A0A1A9XB89 E1ZWA9
Pubmed
19121390
23622113
26354079
26227816
22118469
24438588
+ More
18362917 19820115 24945155 26483478 17510324 20966253 12364791 14747013 17210077 24845553 25348373 25830018 15632085 17994087 28648823 20920257 23761445 22516182 20566863 25401762 26823975 21347285 25244985 29403074 20075255 22936249 10731132 12537572 12537569 27129103 17550304 24495485 24508170 30249741 25315136 26108605 20798317 23537049 21282665 21719571
18362917 19820115 24945155 26483478 17510324 20966253 12364791 14747013 17210077 24845553 25348373 25830018 15632085 17994087 28648823 20920257 23761445 22516182 20566863 25401762 26823975 21347285 25244985 29403074 20075255 22936249 10731132 12537572 12537569 27129103 17550304 24495485 24508170 30249741 25315136 26108605 20798317 23537049 21282665 21719571
EMBL
BABH01014674
NWSH01001431
PCG71248.1
ODYU01003725
SOQ42866.1
GAIX01003675
+ More
JAA88885.1 KQ461155 KPJ08361.1 KQ459603 KPI92984.1 JTDY01000512 KOB76835.1 RSAL01000036 RVE51284.1 AGBW02010202 OWR48977.1 GGFJ01009689 MBW58830.1 AXCM01016726 ATLV01024580 KE525352 KFB51704.1 DS232396 EDS41068.1 GGFL01000611 MBW64789.1 KQ971354 EFA07665.2 GAMD01001315 JAB00276.1 GFDL01007480 JAV27565.1 JXUM01034891 JXUM01034892 JXUM01034893 GAPW01004839 KQ561040 JAC08759.1 KXJ79922.1 GANO01000469 JAB59402.1 CH477833 EAT35814.1 AAAB01008807 EAA04638.3 GGFL01002253 MBW66431.1 KK852527 KDR22021.1 GDHF01032215 JAI20099.1 GAKP01000807 JAC58145.1 OUUW01000001 SPP72732.1 GBXI01000183 JAD14109.1 AJWK01013496 CM000071 EAL26650.2 CH479216 EDW33631.1 NNAY01000618 OXU27368.1 ADMH02000549 ETN65959.1 BT127763 AEE62725.1 DS235430 EEB15627.1 GBHO01007845 GBRD01008447 GDHC01013829 GDHC01001151 JAG35759.1 JAG57374.1 JAQ04800.1 JAQ17478.1 ADTU01011879 PYGN01000302 PSN48922.1 GFDF01011164 JAV02920.1 GFDF01011165 JAV02919.1 AAZX01007897 CH954179 EDV55452.1 APCN01000204 CH480824 EDW56624.1 CM000362 CM002911 EDX08066.1 KMY95532.1 CH963846 EDW72387.1 AE013599 AY061343 GECU01027868 GECU01001350 JAS79838.1 JAT06357.1 GDKW01002915 JAI53680.1 ACPB03015500 GEDC01022559 GEDC01007600 JAS14739.1 JAS29698.1 CM000158 EDW91250.1 CH940648 EDW61924.1 GAMC01001489 JAC05067.1 CH902619 EDV36152.1 KQ978619 KYN29756.1 KQ982934 KYQ49177.1 CH933808 EDW09369.1 GGMS01007018 MBY76221.1 GECL01001452 JAP04672.1 KK107034 QOIP01000008 EZA62050.1 RLU19278.1 GFTR01002667 JAW13759.1 CH916367 EDW00980.1 CP012524 ALC41618.1 JRES01000960 KNC26710.1 GEMB01003624 JAR99594.1 GL445930 EFN88771.1 APGK01019947 APGK01019948 KB740150 ENN81244.1 KB632173 ERL89569.1 KQ981727 KYN37031.1 GFXV01007306 MBW19111.1 ABLF02034513 AK341092 BAH71565.1 KQ414668 KOC64654.1 GL764306 EFZ17846.1 JXJN01022485 KZ288362 PBC26944.1 GL888417 EGI61248.1 GL434791 EFN74513.1
JAA88885.1 KQ461155 KPJ08361.1 KQ459603 KPI92984.1 JTDY01000512 KOB76835.1 RSAL01000036 RVE51284.1 AGBW02010202 OWR48977.1 GGFJ01009689 MBW58830.1 AXCM01016726 ATLV01024580 KE525352 KFB51704.1 DS232396 EDS41068.1 GGFL01000611 MBW64789.1 KQ971354 EFA07665.2 GAMD01001315 JAB00276.1 GFDL01007480 JAV27565.1 JXUM01034891 JXUM01034892 JXUM01034893 GAPW01004839 KQ561040 JAC08759.1 KXJ79922.1 GANO01000469 JAB59402.1 CH477833 EAT35814.1 AAAB01008807 EAA04638.3 GGFL01002253 MBW66431.1 KK852527 KDR22021.1 GDHF01032215 JAI20099.1 GAKP01000807 JAC58145.1 OUUW01000001 SPP72732.1 GBXI01000183 JAD14109.1 AJWK01013496 CM000071 EAL26650.2 CH479216 EDW33631.1 NNAY01000618 OXU27368.1 ADMH02000549 ETN65959.1 BT127763 AEE62725.1 DS235430 EEB15627.1 GBHO01007845 GBRD01008447 GDHC01013829 GDHC01001151 JAG35759.1 JAG57374.1 JAQ04800.1 JAQ17478.1 ADTU01011879 PYGN01000302 PSN48922.1 GFDF01011164 JAV02920.1 GFDF01011165 JAV02919.1 AAZX01007897 CH954179 EDV55452.1 APCN01000204 CH480824 EDW56624.1 CM000362 CM002911 EDX08066.1 KMY95532.1 CH963846 EDW72387.1 AE013599 AY061343 GECU01027868 GECU01001350 JAS79838.1 JAT06357.1 GDKW01002915 JAI53680.1 ACPB03015500 GEDC01022559 GEDC01007600 JAS14739.1 JAS29698.1 CM000158 EDW91250.1 CH940648 EDW61924.1 GAMC01001489 JAC05067.1 CH902619 EDV36152.1 KQ978619 KYN29756.1 KQ982934 KYQ49177.1 CH933808 EDW09369.1 GGMS01007018 MBY76221.1 GECL01001452 JAP04672.1 KK107034 QOIP01000008 EZA62050.1 RLU19278.1 GFTR01002667 JAW13759.1 CH916367 EDW00980.1 CP012524 ALC41618.1 JRES01000960 KNC26710.1 GEMB01003624 JAR99594.1 GL445930 EFN88771.1 APGK01019947 APGK01019948 KB740150 ENN81244.1 KB632173 ERL89569.1 KQ981727 KYN37031.1 GFXV01007306 MBW19111.1 ABLF02034513 AK341092 BAH71565.1 KQ414668 KOC64654.1 GL764306 EFZ17846.1 JXJN01022485 KZ288362 PBC26944.1 GL888417 EGI61248.1 GL434791 EFN74513.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000037510
UP000283053
+ More
UP000007151 UP000075901 UP000075883 UP000030765 UP000075900 UP000002320 UP000075880 UP000007266 UP000069940 UP000249989 UP000008820 UP000075882 UP000007062 UP000027135 UP000268350 UP000092461 UP000001819 UP000008744 UP000075881 UP000075920 UP000075884 UP000215335 UP000075885 UP000000673 UP000009046 UP000069272 UP000005205 UP000076408 UP000245037 UP000192221 UP000002358 UP000008711 UP000075840 UP000001292 UP000000304 UP000007798 UP000000803 UP000015103 UP000002282 UP000008792 UP000007801 UP000192223 UP000078492 UP000075809 UP000009192 UP000053097 UP000279307 UP000005203 UP000001070 UP000092553 UP000079169 UP000095301 UP000037069 UP000008237 UP000019118 UP000030742 UP000078541 UP000007819 UP000095300 UP000091820 UP000053825 UP000078200 UP000092460 UP000242457 UP000007755 UP000092443 UP000000311
UP000007151 UP000075901 UP000075883 UP000030765 UP000075900 UP000002320 UP000075880 UP000007266 UP000069940 UP000249989 UP000008820 UP000075882 UP000007062 UP000027135 UP000268350 UP000092461 UP000001819 UP000008744 UP000075881 UP000075920 UP000075884 UP000215335 UP000075885 UP000000673 UP000009046 UP000069272 UP000005205 UP000076408 UP000245037 UP000192221 UP000002358 UP000008711 UP000075840 UP000001292 UP000000304 UP000007798 UP000000803 UP000015103 UP000002282 UP000008792 UP000007801 UP000192223 UP000078492 UP000075809 UP000009192 UP000053097 UP000279307 UP000005203 UP000001070 UP000092553 UP000079169 UP000095301 UP000037069 UP000008237 UP000019118 UP000030742 UP000078541 UP000007819 UP000095300 UP000091820 UP000053825 UP000078200 UP000092460 UP000242457 UP000007755 UP000092443 UP000000311
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JC05
A0A2A4JHQ4
A0A2H1VQ05
S4PMA3
A0A194QXQ5
A0A194PJS8
+ More
A0A0L7LN27 A0A3S2P3B3 A0A212F5G3 A0A2M4C0I7 A0A182SAP2 A0A182MEI2 A0A084WNB0 A0A182S4P0 B0X5T7 A0A2M4CHQ6 A0A182JB19 D6WRM4 T1DQ81 A0A1Q3FJ65 A0A023EJ15 U5EWP4 Q16N92 A0A1S4FV38 A0A182KRB5 Q7QJF1 A0A2M4CML1 A0A067RDS3 A0A0K8U034 A0A034WVA9 A0A3B0J1K0 A0A0A1XS12 A0A1B0CIK9 Q28WL7 B4H6Z5 A0A182JUH7 A0A182WJ80 A0A182NHV2 A0A232FAA0 A0A182PQE9 W5JTA3 J3JWZ5 E0VQH1 A0A0A9YU71 A0A182FF46 A0A158NCG2 A0A182XYW6 A0A2P8YXF5 A0A1L8D9A6 A0A1L8D8X2 A0A182VHI4 A0A1W4VXS6 K7J455 B3NMV8 A0A182I3B1 A0A182X3Q8 B4I7P9 B4QG30 B4MJU8 Q9W2H1 A0A1B6K4I5 A0A0N7Z8M7 T1HXJ1 A0A1B6DVI9 B4P8Z6 B4LLH2 W8CA92 B3MBY1 A0A1W4W6Q6 A0A151JQW3 A0A151WMZ4 B4KRF3 A0A2S2QEV5 A0A0V0G9B3 A0A026X170 A0A224Y076 A0A087ZV56 B4J6S7 A0A0M5JAA8 A0A1S4E8X3 A0A3Q0ISJ9 A0A1I8MII9 A0A0L0C310 A0A170Y667 E2B678 N6TLB6 U4U6Q3 A0A195F9H6 A0A2H8TZC0 C4WUJ5 A0A1I8NNX6 A0A1A9WJX4 A0A0L7R1D7 E9INI5 A0A1A9V211 A0A1B0BY58 A0A2A3E631 F4WX05 A0A1A9XB89 E1ZWA9
A0A0L7LN27 A0A3S2P3B3 A0A212F5G3 A0A2M4C0I7 A0A182SAP2 A0A182MEI2 A0A084WNB0 A0A182S4P0 B0X5T7 A0A2M4CHQ6 A0A182JB19 D6WRM4 T1DQ81 A0A1Q3FJ65 A0A023EJ15 U5EWP4 Q16N92 A0A1S4FV38 A0A182KRB5 Q7QJF1 A0A2M4CML1 A0A067RDS3 A0A0K8U034 A0A034WVA9 A0A3B0J1K0 A0A0A1XS12 A0A1B0CIK9 Q28WL7 B4H6Z5 A0A182JUH7 A0A182WJ80 A0A182NHV2 A0A232FAA0 A0A182PQE9 W5JTA3 J3JWZ5 E0VQH1 A0A0A9YU71 A0A182FF46 A0A158NCG2 A0A182XYW6 A0A2P8YXF5 A0A1L8D9A6 A0A1L8D8X2 A0A182VHI4 A0A1W4VXS6 K7J455 B3NMV8 A0A182I3B1 A0A182X3Q8 B4I7P9 B4QG30 B4MJU8 Q9W2H1 A0A1B6K4I5 A0A0N7Z8M7 T1HXJ1 A0A1B6DVI9 B4P8Z6 B4LLH2 W8CA92 B3MBY1 A0A1W4W6Q6 A0A151JQW3 A0A151WMZ4 B4KRF3 A0A2S2QEV5 A0A0V0G9B3 A0A026X170 A0A224Y076 A0A087ZV56 B4J6S7 A0A0M5JAA8 A0A1S4E8X3 A0A3Q0ISJ9 A0A1I8MII9 A0A0L0C310 A0A170Y667 E2B678 N6TLB6 U4U6Q3 A0A195F9H6 A0A2H8TZC0 C4WUJ5 A0A1I8NNX6 A0A1A9WJX4 A0A0L7R1D7 E9INI5 A0A1A9V211 A0A1B0BY58 A0A2A3E631 F4WX05 A0A1A9XB89 E1ZWA9
Ontologies
Topology
Subcellular location
Membrane
SignalP
Position: 1 - 23,
Likelihood: 0.998580
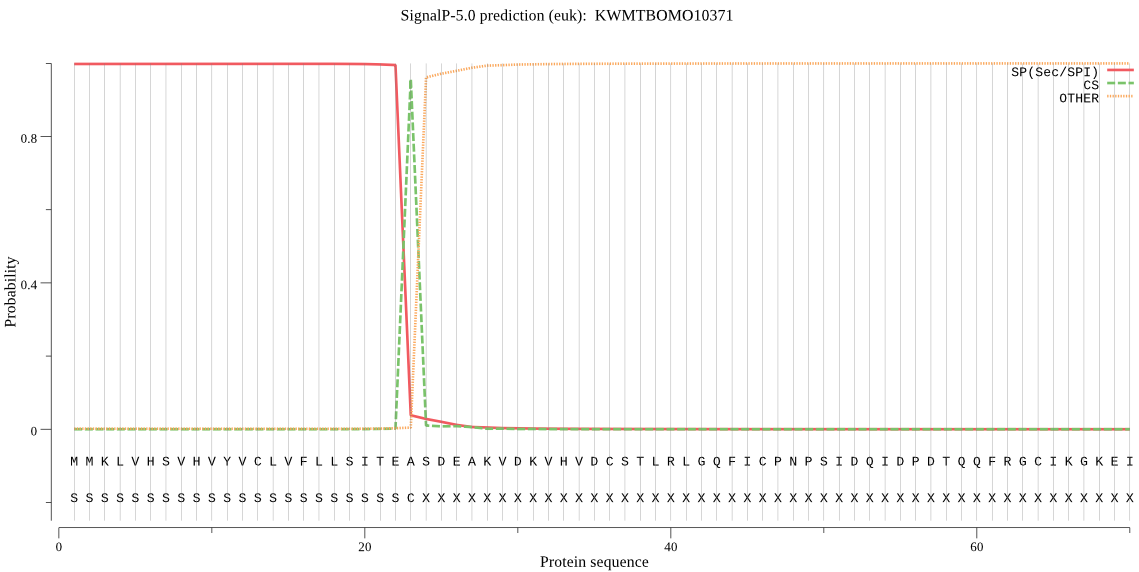
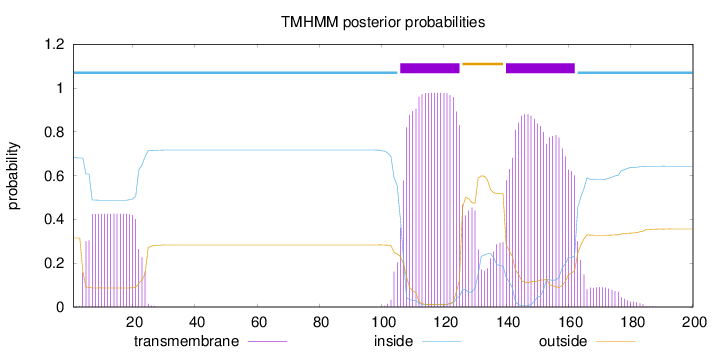
Length:
200
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
49.68894
Exp number, first 60 AAs:
7.79454
Total prob of N-in:
0.68436
inside
1 - 105
TMhelix
106 - 125
outside
126 - 139
TMhelix
140 - 162
inside
163 - 200
Population Genetic Test Statistics
Pi
187.412605
Theta
193.28972
Tajima's D
-0.812993
CLR
2.293022
CSRT
0.176291185440728
Interpretation
Uncertain
