Gene
KWMTBOMO10370
Pre Gene Modal
BGIBMGA007060
Annotation
PREDICTED:_evolutionarily_conserved_signaling_intermediate_in_Toll_pathway?_mitochondrial_[Papilio_xuthus]
Full name
Evolutionarily conserved signaling intermediate in Toll pathway, mitochondrial
Location in the cell
Cytoplasmic Reliability : 2.341
Sequence
CDS
ATGTTTGAAAATCAGGACAACAGAAGAAGAGGACACGTCGAATTTATCTATGCCGCCTTAGCTAGAATGAAGGAATTTGGTGTGCATAAAGACTTACAAGCGTACAAAGCATTAGTAGACGTACTGCCCAAAGGCAAATTTATACCGTCAAATATATTTCAGGCTGAATTCATGCACTATCCGAAGCAACAGCAGTGCGCTGTGGATCTCCTTGAACAGATGGAAGATAATAAAGTAATGCCAGACAGTGAACTAGAGCAAATGTTATTGAACGTTTTTGGAAAAAGAGGCATACCTCTTAGAAAGTTTTGGCGCATGCTGTACTGGATGCCTAAATTCAAGAATCTAAGTCCATGGTATTTACCAGATGAATTGCCGAATGACACATTAGAACTTGCCAAAATGGCTATTCATCGGATAACTTCCGTTGATCCTACTATGAAAATTGAAATTTGGCAGACAGAAGAAATTGAAGCTTGCATAGATAAAACATGGATAGTAAGTGCCCAAAGTGAAACACAAAAGATATTACTCTCAGAGCAACCCCCTGATGAGCCCCTTGTTGTAAGAGGACCTTACGAAGTGTATTTAAAGGAGCAAGTTGTCACTTACTTTACATTATTGGGTAAAGTAAGACCAGAACATAAAGATGACTTTGACCAAGATGATGTATCAAATTTAAAGAAACCTTCCGGAATACCTGGTTTTATTGGTCAAACTACATTACCCGTAGTTAAATGTACTGTACATGAGCAGGATGATGGTACAATAGTAGCTGTATGTGCCACAGGTTCATCATCACGAGATTCTTTGTTATCGTGGATAAGATTATTAGAGAAATATGACAATTCAGTTCTATCAAATCTACCAGTGATTTTCACTCTTACTGCACCTAGCAATGACGTTGTAGTACAGGAAACACAGCCAGCTGCCGAAGATGTAAAAAGAGTGCAAAATTAA
Protein
MFENQDNRRRGHVEFIYAALARMKEFGVHKDLQAYKALVDVLPKGKFIPSNIFQAEFMHYPKQQQCAVDLLEQMEDNKVMPDSELEQMLLNVFGKRGIPLRKFWRMLYWMPKFKNLSPWYLPDELPNDTLELAKMAIHRITSVDPTMKIEIWQTEEIEACIDKTWIVSAQSETQKILLSEQPPDEPLVVRGPYEVYLKEQVVTYFTLLGKVRPEHKDDFDQDDVSNLKKPSGIPGFIGQTTLPVVKCTVHEQDDGTIVAVCATGSSSRDSLLSWIRLLEKYDNSVLSNLPVIFTLTAPSNDVVVQETQPAAEDVKRVQN
Summary
Description
Involved in the innate immune response. Promotes the production of antibacterial peptides.
Required for efficient assembly of mitochondrial NADH:ubiquinone oxidoreductase.
Required for efficient assembly of mitochondrial NADH:ubiquinone oxidoreductase.
Subunit
Interacts with Traf6.
Similarity
Belongs to the ECSIT family.
Keywords
Complete proteome
Cytoplasm
Immunity
Innate immunity
Mitochondrion
Nucleus
Reference proteome
Transit peptide
Feature
chain Evolutionarily conserved signaling intermediate in Toll pathway, mitochondrial
Uniprot
A0A194PIV7
H9JC15
A0A194QTN2
A0A0L7LNA1
A0A2H1VPY2
A0A2A4JHG0
+ More
A0A1E1W383 A0A346RAD4 A0A212F5G8 A0A1E1W3E6 A0A1W4XFH4 A0A1Y1N150 A0A1Y1MVW3 T1IF30 W5JEM5 D6WU07 A0A182KVX2 A0A182FCZ7 A0A023F2Q5 A0A069DSZ5 A0A182ICX4 A0A182UBW7 Q7QI51 A0A182J825 A0A182WRZ5 A0A182NA29 A0A0V0G7N9 A0A182WK42 A0A2M4AU66 A0A182UQ16 A0A2M4ASX8 A0A182LYP6 A0A182JW46 A0A182RQD7 A0A182SRV3 A0A1J1IRF5 A0A182YP07 A0A084VXL8 A0A182QH61 A0A182HD34 B4K710 Q17B53 A0A067QJH9 A0A034VIQ5 A0A1W4V3Y1 A0A2J7QED4 A0A0K8U9F0 B4MC61 A0A1B0G2E3 A0A0A9XBY9 B3NYI6 B0W9N2 B4I491 W8CCK7 B4QXL9 B6IDJ7 Q9U6M0 B4NHG1 A0A1B6C3D7 D3TPK5 B4PVJ6 A0A1A9WPG9 Q297W7 A0A336MM82 A0A1G4P228 B3LWB9 A0A1B6K3M5 A0A3B0JHR2 A0A0M3QXN1 A0A1B6FQ69 A0A1B6I5L3 A0A0P4WDQ0 A0A1A9ZJH5 A0A1I8NE33 A0A097F5U1 A0A1A9XTD6 A0A1B0BZR9 B4JGQ9 U4UCT0 A0A0T6B347 J3JWY6 N6TV89 A0A1I8PEW7 A0A0L0BXT1 A0A1B6HR21 A0A1Q3F8Y8 B4G354 A0A0N8B818 A0A0P5J8T7 A0A0P4WI97 A0A0P5GHQ6 E9HFW4 A0A1D2MR97 A0A2R7WPM6 A0A0P5W4X3 E2APA7 A0A0P5F948 A0A2S2Q7B6 A0A0J7L406 E9ICX0
A0A1E1W383 A0A346RAD4 A0A212F5G8 A0A1E1W3E6 A0A1W4XFH4 A0A1Y1N150 A0A1Y1MVW3 T1IF30 W5JEM5 D6WU07 A0A182KVX2 A0A182FCZ7 A0A023F2Q5 A0A069DSZ5 A0A182ICX4 A0A182UBW7 Q7QI51 A0A182J825 A0A182WRZ5 A0A182NA29 A0A0V0G7N9 A0A182WK42 A0A2M4AU66 A0A182UQ16 A0A2M4ASX8 A0A182LYP6 A0A182JW46 A0A182RQD7 A0A182SRV3 A0A1J1IRF5 A0A182YP07 A0A084VXL8 A0A182QH61 A0A182HD34 B4K710 Q17B53 A0A067QJH9 A0A034VIQ5 A0A1W4V3Y1 A0A2J7QED4 A0A0K8U9F0 B4MC61 A0A1B0G2E3 A0A0A9XBY9 B3NYI6 B0W9N2 B4I491 W8CCK7 B4QXL9 B6IDJ7 Q9U6M0 B4NHG1 A0A1B6C3D7 D3TPK5 B4PVJ6 A0A1A9WPG9 Q297W7 A0A336MM82 A0A1G4P228 B3LWB9 A0A1B6K3M5 A0A3B0JHR2 A0A0M3QXN1 A0A1B6FQ69 A0A1B6I5L3 A0A0P4WDQ0 A0A1A9ZJH5 A0A1I8NE33 A0A097F5U1 A0A1A9XTD6 A0A1B0BZR9 B4JGQ9 U4UCT0 A0A0T6B347 J3JWY6 N6TV89 A0A1I8PEW7 A0A0L0BXT1 A0A1B6HR21 A0A1Q3F8Y8 B4G354 A0A0N8B818 A0A0P5J8T7 A0A0P4WI97 A0A0P5GHQ6 E9HFW4 A0A1D2MR97 A0A2R7WPM6 A0A0P5W4X3 E2APA7 A0A0P5F948 A0A2S2Q7B6 A0A0J7L406 E9ICX0
Pubmed
26354079
19121390
26227816
29910977
22118469
28004739
+ More
20920257 23761445 18362917 19820115 20966253 25474469 26334808 12364791 14747013 17210077 25244985 24438588 26483478 17994087 17510324 24845553 25348373 25401762 24495485 10465784 10731132 12537572 12537569 20353571 17550304 15632085 25315136 24796866 23537049 22516182 26108605 21292972 27289101 20798317 21282665
20920257 23761445 18362917 19820115 20966253 25474469 26334808 12364791 14747013 17210077 25244985 24438588 26483478 17994087 17510324 24845553 25348373 25401762 24495485 10465784 10731132 12537572 12537569 20353571 17550304 15632085 25315136 24796866 23537049 22516182 26108605 21292972 27289101 20798317 21282665
EMBL
KQ459603
KPI92983.1
BABH01014674
KQ461155
KPJ08360.1
JTDY01000512
+ More
KOB76834.1 ODYU01003725 SOQ42867.1 NWSH01001431 PCG71249.1 GDQN01009611 JAT81443.1 MH200924 AXS59134.1 AGBW02010202 OWR48978.1 GDQN01009554 JAT81500.1 GEZM01020452 JAV89267.1 GEZM01020451 JAV89268.1 ACPB03009564 ADMH02001370 ETN62807.1 KQ971352 EFA07356.1 GBBI01003102 JAC15610.1 GBGD01002053 JAC86836.1 APCN01006058 AAAB01008810 EAA04813.4 GECL01002054 JAP04070.1 GGFK01011024 MBW44345.1 GGFK01010584 MBW43905.1 AXCM01001615 CVRI01000055 CRL01097.1 ATLV01018097 KE525212 KFB42712.1 AXCN02001953 JXUM01005146 KQ560187 KXJ83937.1 CH933806 EDW15297.2 CH477325 EAT43515.1 KK853286 KDR08946.1 GAKP01017514 JAC41438.1 NEVH01015316 PNF26923.1 GDHF01029055 JAI23259.1 CH940656 EDW58682.2 CCAG010003931 GBHO01026090 GBRD01017769 JAG17514.1 JAG48058.1 CH954181 EDV47967.1 DS231865 EDS40351.1 CH480821 EDW55034.1 GAMC01003526 JAC03030.1 CM000364 EDX11805.1 BT050437 ACJ13144.1 AF182511 AE014297 AY070908 CH964272 EDW84637.1 GEDC01029277 JAS08021.1 EZ423357 ADD19633.1 CM000160 EDW97805.1 CM000070 EAL28088.2 UFQT01001591 SSX31090.1 LT626243 SCW25224.1 CH902617 EDV43752.1 GECU01001645 JAT06062.1 OUUW01000005 SPP80243.1 CP012526 ALC46159.1 GECZ01017422 JAS52347.1 GECU01025521 JAS82185.1 GDRN01097353 GDRN01097352 GDRN01097350 JAI59131.1 KF995355 AIT17846.1 JXJN01023274 CH916369 EDV92663.1 KB631330 KB632210 ERL84242.1 ERL90133.1 LJIG01016030 KRT81824.1 BT127754 AEE62716.1 APGK01050708 APGK01057170 KB741277 KB741176 ENN71316.1 ENN73190.1 JRES01001168 KNC24858.1 GECU01030605 JAS77101.1 GFDL01011058 JAV23987.1 CH479179 EDW24249.1 GDIQ01203715 JAK48010.1 GDIQ01209414 JAK42311.1 GDIP01254874 JAI68527.1 GDIQ01246644 JAK05081.1 GL732637 EFX69389.1 LJIJ01000677 ODM95421.1 KK855234 PTY21572.1 GDIP01091506 JAM12209.1 GL441497 EFN64741.1 GDIQ01258268 JAJ93456.1 GGMS01004425 MBY73628.1 LBMM01000921 KMQ97209.1 GL762419 EFZ21583.1
KOB76834.1 ODYU01003725 SOQ42867.1 NWSH01001431 PCG71249.1 GDQN01009611 JAT81443.1 MH200924 AXS59134.1 AGBW02010202 OWR48978.1 GDQN01009554 JAT81500.1 GEZM01020452 JAV89267.1 GEZM01020451 JAV89268.1 ACPB03009564 ADMH02001370 ETN62807.1 KQ971352 EFA07356.1 GBBI01003102 JAC15610.1 GBGD01002053 JAC86836.1 APCN01006058 AAAB01008810 EAA04813.4 GECL01002054 JAP04070.1 GGFK01011024 MBW44345.1 GGFK01010584 MBW43905.1 AXCM01001615 CVRI01000055 CRL01097.1 ATLV01018097 KE525212 KFB42712.1 AXCN02001953 JXUM01005146 KQ560187 KXJ83937.1 CH933806 EDW15297.2 CH477325 EAT43515.1 KK853286 KDR08946.1 GAKP01017514 JAC41438.1 NEVH01015316 PNF26923.1 GDHF01029055 JAI23259.1 CH940656 EDW58682.2 CCAG010003931 GBHO01026090 GBRD01017769 JAG17514.1 JAG48058.1 CH954181 EDV47967.1 DS231865 EDS40351.1 CH480821 EDW55034.1 GAMC01003526 JAC03030.1 CM000364 EDX11805.1 BT050437 ACJ13144.1 AF182511 AE014297 AY070908 CH964272 EDW84637.1 GEDC01029277 JAS08021.1 EZ423357 ADD19633.1 CM000160 EDW97805.1 CM000070 EAL28088.2 UFQT01001591 SSX31090.1 LT626243 SCW25224.1 CH902617 EDV43752.1 GECU01001645 JAT06062.1 OUUW01000005 SPP80243.1 CP012526 ALC46159.1 GECZ01017422 JAS52347.1 GECU01025521 JAS82185.1 GDRN01097353 GDRN01097352 GDRN01097350 JAI59131.1 KF995355 AIT17846.1 JXJN01023274 CH916369 EDV92663.1 KB631330 KB632210 ERL84242.1 ERL90133.1 LJIG01016030 KRT81824.1 BT127754 AEE62716.1 APGK01050708 APGK01057170 KB741277 KB741176 ENN71316.1 ENN73190.1 JRES01001168 KNC24858.1 GECU01030605 JAS77101.1 GFDL01011058 JAV23987.1 CH479179 EDW24249.1 GDIQ01203715 JAK48010.1 GDIQ01209414 JAK42311.1 GDIP01254874 JAI68527.1 GDIQ01246644 JAK05081.1 GL732637 EFX69389.1 LJIJ01000677 ODM95421.1 KK855234 PTY21572.1 GDIP01091506 JAM12209.1 GL441497 EFN64741.1 GDIQ01258268 JAJ93456.1 GGMS01004425 MBY73628.1 LBMM01000921 KMQ97209.1 GL762419 EFZ21583.1
Proteomes
UP000053268
UP000005204
UP000053240
UP000037510
UP000218220
UP000007151
+ More
UP000192223 UP000015103 UP000000673 UP000007266 UP000075882 UP000069272 UP000075840 UP000075902 UP000007062 UP000075880 UP000076407 UP000075884 UP000075920 UP000075903 UP000075883 UP000075881 UP000075900 UP000075901 UP000183832 UP000076408 UP000030765 UP000075886 UP000069940 UP000249989 UP000009192 UP000008820 UP000027135 UP000192221 UP000235965 UP000008792 UP000092444 UP000008711 UP000002320 UP000001292 UP000000304 UP000000803 UP000007798 UP000002282 UP000091820 UP000001819 UP000007801 UP000268350 UP000092553 UP000092445 UP000095301 UP000092443 UP000092460 UP000001070 UP000030742 UP000019118 UP000095300 UP000037069 UP000008744 UP000000305 UP000094527 UP000000311 UP000036403
UP000192223 UP000015103 UP000000673 UP000007266 UP000075882 UP000069272 UP000075840 UP000075902 UP000007062 UP000075880 UP000076407 UP000075884 UP000075920 UP000075903 UP000075883 UP000075881 UP000075900 UP000075901 UP000183832 UP000076408 UP000030765 UP000075886 UP000069940 UP000249989 UP000009192 UP000008820 UP000027135 UP000192221 UP000235965 UP000008792 UP000092444 UP000008711 UP000002320 UP000001292 UP000000304 UP000000803 UP000007798 UP000002282 UP000091820 UP000001819 UP000007801 UP000268350 UP000092553 UP000092445 UP000095301 UP000092443 UP000092460 UP000001070 UP000030742 UP000019118 UP000095300 UP000037069 UP000008744 UP000000305 UP000094527 UP000000311 UP000036403
Interpro
Gene 3D
ProteinModelPortal
A0A194PIV7
H9JC15
A0A194QTN2
A0A0L7LNA1
A0A2H1VPY2
A0A2A4JHG0
+ More
A0A1E1W383 A0A346RAD4 A0A212F5G8 A0A1E1W3E6 A0A1W4XFH4 A0A1Y1N150 A0A1Y1MVW3 T1IF30 W5JEM5 D6WU07 A0A182KVX2 A0A182FCZ7 A0A023F2Q5 A0A069DSZ5 A0A182ICX4 A0A182UBW7 Q7QI51 A0A182J825 A0A182WRZ5 A0A182NA29 A0A0V0G7N9 A0A182WK42 A0A2M4AU66 A0A182UQ16 A0A2M4ASX8 A0A182LYP6 A0A182JW46 A0A182RQD7 A0A182SRV3 A0A1J1IRF5 A0A182YP07 A0A084VXL8 A0A182QH61 A0A182HD34 B4K710 Q17B53 A0A067QJH9 A0A034VIQ5 A0A1W4V3Y1 A0A2J7QED4 A0A0K8U9F0 B4MC61 A0A1B0G2E3 A0A0A9XBY9 B3NYI6 B0W9N2 B4I491 W8CCK7 B4QXL9 B6IDJ7 Q9U6M0 B4NHG1 A0A1B6C3D7 D3TPK5 B4PVJ6 A0A1A9WPG9 Q297W7 A0A336MM82 A0A1G4P228 B3LWB9 A0A1B6K3M5 A0A3B0JHR2 A0A0M3QXN1 A0A1B6FQ69 A0A1B6I5L3 A0A0P4WDQ0 A0A1A9ZJH5 A0A1I8NE33 A0A097F5U1 A0A1A9XTD6 A0A1B0BZR9 B4JGQ9 U4UCT0 A0A0T6B347 J3JWY6 N6TV89 A0A1I8PEW7 A0A0L0BXT1 A0A1B6HR21 A0A1Q3F8Y8 B4G354 A0A0N8B818 A0A0P5J8T7 A0A0P4WI97 A0A0P5GHQ6 E9HFW4 A0A1D2MR97 A0A2R7WPM6 A0A0P5W4X3 E2APA7 A0A0P5F948 A0A2S2Q7B6 A0A0J7L406 E9ICX0
A0A1E1W383 A0A346RAD4 A0A212F5G8 A0A1E1W3E6 A0A1W4XFH4 A0A1Y1N150 A0A1Y1MVW3 T1IF30 W5JEM5 D6WU07 A0A182KVX2 A0A182FCZ7 A0A023F2Q5 A0A069DSZ5 A0A182ICX4 A0A182UBW7 Q7QI51 A0A182J825 A0A182WRZ5 A0A182NA29 A0A0V0G7N9 A0A182WK42 A0A2M4AU66 A0A182UQ16 A0A2M4ASX8 A0A182LYP6 A0A182JW46 A0A182RQD7 A0A182SRV3 A0A1J1IRF5 A0A182YP07 A0A084VXL8 A0A182QH61 A0A182HD34 B4K710 Q17B53 A0A067QJH9 A0A034VIQ5 A0A1W4V3Y1 A0A2J7QED4 A0A0K8U9F0 B4MC61 A0A1B0G2E3 A0A0A9XBY9 B3NYI6 B0W9N2 B4I491 W8CCK7 B4QXL9 B6IDJ7 Q9U6M0 B4NHG1 A0A1B6C3D7 D3TPK5 B4PVJ6 A0A1A9WPG9 Q297W7 A0A336MM82 A0A1G4P228 B3LWB9 A0A1B6K3M5 A0A3B0JHR2 A0A0M3QXN1 A0A1B6FQ69 A0A1B6I5L3 A0A0P4WDQ0 A0A1A9ZJH5 A0A1I8NE33 A0A097F5U1 A0A1A9XTD6 A0A1B0BZR9 B4JGQ9 U4UCT0 A0A0T6B347 J3JWY6 N6TV89 A0A1I8PEW7 A0A0L0BXT1 A0A1B6HR21 A0A1Q3F8Y8 B4G354 A0A0N8B818 A0A0P5J8T7 A0A0P4WI97 A0A0P5GHQ6 E9HFW4 A0A1D2MR97 A0A2R7WPM6 A0A0P5W4X3 E2APA7 A0A0P5F948 A0A2S2Q7B6 A0A0J7L406 E9ICX0
Ontologies
PANTHER
Topology
Subcellular location
Cytoplasm
Nucleus
Mitochondrion
Nucleus
Mitochondrion
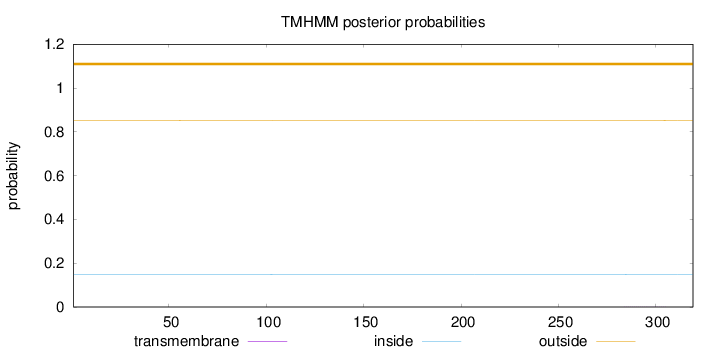
Length:
319
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01117
Exp number, first 60 AAs:
0.00114
Total prob of N-in:
0.14909
outside
1 - 319
Population Genetic Test Statistics
Pi
234.78729
Theta
237.813733
Tajima's D
-0.565614
CLR
0
CSRT
0.231588420578971
Interpretation
Uncertain
