Gene
KWMTBOMO10366
Pre Gene Modal
BGIBMGA007058
Annotation
putative_tubulin_polyglutamylase_TTLL1_[Papilio_machaon]
Location in the cell
Cytoplasmic Reliability : 1.683 Nuclear Reliability : 1.978
Sequence
CDS
ATGGAAAAGGGAAAAGTAACTTATTGCACAGATTTGGAGAAATCTGTGATCATCAGCAACTTCGAGCGACGAGGATGGGTCCAGGTAGGCCCAGAAGATGAATGGAATTTTTACTGGTCTTTCACTCAAAACTGCCGCAACATTTTTAGTATCGAAAGTGGGTACAGAATGAACGACAACCAAATGATAAATCACTTCCCTAACCACTACGAGCTGTCTCGTAAAGATTTATTAGTAAAAAATATCAAAAGATACAGAAAAGAACTTGAAAGGGAAGGCAATCCTTTAGCTGAAAAAACGGATTTGGTGCTCCCAAGCGGGCAAGTTGTCTCGCGATATTTACATTTAGATTTTATACCAGTCACTTACGTTTTGCCTGCCGACTACAATATGTTTGTCGAAGAATACAGAAAGTCTCCGCAAAGTACGTGGATCATGAAGCCTTGTGGAAAATCTCAAGGGGCGGGAATTTTCTTGATCAATAAACTTTCCAAACTGAAAAAGTGGTCGCGGGAAGCGAAAACTCCTTTCCATCCTCAACTCAGCAACAAAGAAAGTTATGTAATATCTCGATACATCGACAACCCTCTTTTGATTGGCGGCAAGAAATTTGATCTGAGACTTTATGTACTCGTGACATCGTTTCGACCACTGAAAGCGTATCTGTTTCGACATGGCTTCTGCAGATTTTGTACAGTGAAATATGACACTAGCGTTACTGAACTCGATAACATGTACGTGCATTTAACGAATGTCAGCGTTCAAAAACACGGAGGGGATTACAACAGCTTACACGGCGGGAAAATGAGTATTCAGAATTTTCGCTTTTATTTAGAAGGGACTAGAGGACGGGCTGTTACCGAAAAGCTATTTGCAGACATGCAATGGTTGATTGTTCATTCACTCAAAGCGGTAGCTCCCGTAATGGCCAATGATAGGCATTGCTTCGAATGCTATGGCTACGACATTATAATTGACAATACTCTGAAACCTTGGCTAGTGGAAGTAAACGCTTCGCCATCTCTGCAATCGACGACTCATAATGACAGAATACTAAAGTACAAATTGATTGATAATATTTTCTCAGTTGTCGTGCCCCCTGACGGGATTCCCGATGCTAGATGGAATAAAATACCTACTACCGAAGCGCTTGGAGACTTTGATCTACTAATCGATGAAGACCTTATGGAAAAAGAAGACATGAATACGCGTTTTTGTAAATTTCAACGAGTCAAGCAATAA
Protein
MEKGKVTYCTDLEKSVIISNFERRGWVQVGPEDEWNFYWSFTQNCRNIFSIESGYRMNDNQMINHFPNHYELSRKDLLVKNIKRYRKELEREGNPLAEKTDLVLPSGQVVSRYLHLDFIPVTYVLPADYNMFVEEYRKSPQSTWIMKPCGKSQGAGIFLINKLSKLKKWSREAKTPFHPQLSNKESYVISRYIDNPLLIGGKKFDLRLYVLVTSFRPLKAYLFRHGFCRFCTVKYDTSVTELDNMYVHLTNVSVQKHGGDYNSLHGGKMSIQNFRFYLEGTRGRAVTEKLFADMQWLIVHSLKAVAPVMANDRHCFECYGYDIIIDNTLKPWLVEVNASPSLQSTTHNDRILKYKLIDNIFSVVVPPDGIPDARWNKIPTTEALGDFDLLIDEDLMEKEDMNTRFCKFQRVKQ
Summary
Uniprot
H9JC13
A0A2A4K671
A0A2H1VZ45
A0A212FK33
A0A0N1IA08
A0A0L7KNZ4
+ More
A0A194QBN7 A0A0N1PIY6 H9JC14 A0A2A4K6G2 A0A194QCX3 A0A0L7L178 A0A2H1VYQ9 A0A1I8NH69 A0A336M6P3 A0A336KXR1 A0A182YE16 A0A232ETA9 A0A1I8PJ47 A0A0L0C9N8 A0A182R930 K7IYY3 A0A0M8ZWN3 A0A151I174 E2B7T0 A0A158NKC0 A0A0A1XII5 A0A182W857 W8C190 A0A0K8WGN2 A0A151WS30 A0A034WF49 A0A182JV02 A0A182P7N6 W5JMM9 A0A182QW01 A0A182IM81 A0A1S4H3P2 A0A182HIW0 A0A1B0GHE9 A0A084VJW0 Q17AY7 A0A182NQ30 Q7QDP1 A0A182GLP6 A0A3F2Z218 A0A182FZS6 A0A1A9W342 A0A195EIM7 A0A2A3E6H8 B0WZV2 A0A087ZY45 A0A310SCC6 A0A2J7PMN2 A0A195FBA5 A0A1A9VBY4 A0A1A9YDG0 A0A1B0G1Z0 B4KXA5 B3M9J3 A0A1W4VNM4 B4PIA7 B4MMY1 B4QR13 A0A0M3QVV4 B3NG56 Q9VZ97 Q29EA9 B4HU95 A0A1B6K0G9 B4H1S0 A0A1B6EK26 A0A3B0J736 B4IZ74 A0A1A9WDU0 B4MFZ2 D2A295 A0A2A4K5S3 A0A0L7KP30 A0A154P731 A0A182LGU7 U4U8A8 N6TVJ1 A0A026WFF6 A0A2H1VYQ7 A0A1W4XH13 A0A1J1HTR7 A0A067RI56 A0A1Y1LX05 A0A2R7VTP4 A0A1B0DCH7 A0A224XPD9 A0A1B0FAE4 A0A3L8DKS2 E0VPT2 A0A2P8XK43 H9JC09 A0A2S2NY11 J9JN31 A0A2H8TGB9
A0A194QBN7 A0A0N1PIY6 H9JC14 A0A2A4K6G2 A0A194QCX3 A0A0L7L178 A0A2H1VYQ9 A0A1I8NH69 A0A336M6P3 A0A336KXR1 A0A182YE16 A0A232ETA9 A0A1I8PJ47 A0A0L0C9N8 A0A182R930 K7IYY3 A0A0M8ZWN3 A0A151I174 E2B7T0 A0A158NKC0 A0A0A1XII5 A0A182W857 W8C190 A0A0K8WGN2 A0A151WS30 A0A034WF49 A0A182JV02 A0A182P7N6 W5JMM9 A0A182QW01 A0A182IM81 A0A1S4H3P2 A0A182HIW0 A0A1B0GHE9 A0A084VJW0 Q17AY7 A0A182NQ30 Q7QDP1 A0A182GLP6 A0A3F2Z218 A0A182FZS6 A0A1A9W342 A0A195EIM7 A0A2A3E6H8 B0WZV2 A0A087ZY45 A0A310SCC6 A0A2J7PMN2 A0A195FBA5 A0A1A9VBY4 A0A1A9YDG0 A0A1B0G1Z0 B4KXA5 B3M9J3 A0A1W4VNM4 B4PIA7 B4MMY1 B4QR13 A0A0M3QVV4 B3NG56 Q9VZ97 Q29EA9 B4HU95 A0A1B6K0G9 B4H1S0 A0A1B6EK26 A0A3B0J736 B4IZ74 A0A1A9WDU0 B4MFZ2 D2A295 A0A2A4K5S3 A0A0L7KP30 A0A154P731 A0A182LGU7 U4U8A8 N6TVJ1 A0A026WFF6 A0A2H1VYQ7 A0A1W4XH13 A0A1J1HTR7 A0A067RI56 A0A1Y1LX05 A0A2R7VTP4 A0A1B0DCH7 A0A224XPD9 A0A1B0FAE4 A0A3L8DKS2 E0VPT2 A0A2P8XK43 H9JC09 A0A2S2NY11 J9JN31 A0A2H8TGB9
Pubmed
19121390
22118469
26354079
26227816
25315136
25244985
+ More
28648823 26108605 20075255 20798317 21347285 25830018 24495485 25348373 20920257 23761445 12364791 24438588 17510324 26483478 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 18057021 18362917 19820115 20966253 23537049 24508170 24845553 28004739 30249741 20566863 29403074
28648823 26108605 20075255 20798317 21347285 25830018 24495485 25348373 20920257 23761445 12364791 24438588 17510324 26483478 17994087 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 18057021 18362917 19820115 20966253 23537049 24508170 24845553 28004739 30249741 20566863 29403074
EMBL
BABH01014644
NWSH01000129
PCG79263.1
ODYU01005278
SOQ45976.1
AGBW02008157
+ More
OWR54091.1 KQ459998 KPJ18501.1 JTDY01007972 KOB64825.1 KQ459232 KPJ02829.1 KPJ18500.1 BABH01014657 PCG79262.1 KPJ02830.1 JTDY01003629 KOB69242.1 SOQ45979.1 UFQS01000205 UFQT01000205 SSX01318.1 SSX21698.1 UFQS01001174 UFQT01001174 SSX09405.1 SSX29307.1 NNAY01002297 OXU21595.1 JRES01000720 KNC28966.1 KQ435847 KOX71133.1 KQ976588 KYM79691.1 GL446209 EFN88223.1 ADTU01018768 GBXI01003562 JAD10730.1 GAMC01003642 JAC02914.1 GDHF01002077 JAI50237.1 KQ982782 KYQ50719.1 GAKP01006544 JAC52408.1 ADMH02000671 ETN65381.1 AXCN02002267 AAAB01008849 APCN01006664 APCN01006665 AJWK01004543 ATLV01013905 KE524908 KFB38254.1 CH477328 EAT43427.1 EAA07118.4 JXUM01016072 JXUM01016073 KQ560449 KXJ82367.1 KQ978881 KYN27752.1 KZ288369 PBC26769.1 DS232218 EDS37777.1 KQ769179 OAD52913.1 NEVH01023972 PNF17591.1 KQ981693 KYN37698.1 CCAG010017015 CH933809 EDW17563.1 CH902618 EDV38999.2 CM000159 EDW93449.1 CH963847 EDW73537.1 CM000363 CM002912 EDX09250.1 KMY97661.1 CP012525 ALC43049.1 CH954178 EDV50957.1 AE014296 BT025844 AAF47928.1 ABF85744.1 CH379070 EAL30152.2 CH480817 EDW50516.1 GECU01002761 JAT04946.1 CH479203 EDW30272.1 GECZ01031518 JAS38251.1 OUUW01000002 SPP77575.1 CH917938 CH916366 EDV90440.1 EDV95596.1 CH940669 EDW58253.1 KRF78201.1 KQ971338 EFA02053.1 PCG79264.1 KOB64826.1 KQ434829 KZC07739.1 KB632081 ERL88578.1 APGK01053111 APGK01053112 KB741221 ENN72416.1 KK107238 EZA54792.1 SOQ45975.1 CVRI01000020 CRK91387.1 KK852498 KDR22678.1 GEZM01044876 JAV78072.1 KK854082 PTY10893.1 AJVK01005093 AJVK01005094 GFTR01006004 JAW10422.1 CCAG010017010 QOIP01000007 RLU21017.1 DS235371 EEB15388.1 PYGN01001874 PSN32376.1 GGMR01009451 MBY22070.1 ABLF02036147 GFXV01001361 MBW13166.1
OWR54091.1 KQ459998 KPJ18501.1 JTDY01007972 KOB64825.1 KQ459232 KPJ02829.1 KPJ18500.1 BABH01014657 PCG79262.1 KPJ02830.1 JTDY01003629 KOB69242.1 SOQ45979.1 UFQS01000205 UFQT01000205 SSX01318.1 SSX21698.1 UFQS01001174 UFQT01001174 SSX09405.1 SSX29307.1 NNAY01002297 OXU21595.1 JRES01000720 KNC28966.1 KQ435847 KOX71133.1 KQ976588 KYM79691.1 GL446209 EFN88223.1 ADTU01018768 GBXI01003562 JAD10730.1 GAMC01003642 JAC02914.1 GDHF01002077 JAI50237.1 KQ982782 KYQ50719.1 GAKP01006544 JAC52408.1 ADMH02000671 ETN65381.1 AXCN02002267 AAAB01008849 APCN01006664 APCN01006665 AJWK01004543 ATLV01013905 KE524908 KFB38254.1 CH477328 EAT43427.1 EAA07118.4 JXUM01016072 JXUM01016073 KQ560449 KXJ82367.1 KQ978881 KYN27752.1 KZ288369 PBC26769.1 DS232218 EDS37777.1 KQ769179 OAD52913.1 NEVH01023972 PNF17591.1 KQ981693 KYN37698.1 CCAG010017015 CH933809 EDW17563.1 CH902618 EDV38999.2 CM000159 EDW93449.1 CH963847 EDW73537.1 CM000363 CM002912 EDX09250.1 KMY97661.1 CP012525 ALC43049.1 CH954178 EDV50957.1 AE014296 BT025844 AAF47928.1 ABF85744.1 CH379070 EAL30152.2 CH480817 EDW50516.1 GECU01002761 JAT04946.1 CH479203 EDW30272.1 GECZ01031518 JAS38251.1 OUUW01000002 SPP77575.1 CH917938 CH916366 EDV90440.1 EDV95596.1 CH940669 EDW58253.1 KRF78201.1 KQ971338 EFA02053.1 PCG79264.1 KOB64826.1 KQ434829 KZC07739.1 KB632081 ERL88578.1 APGK01053111 APGK01053112 KB741221 ENN72416.1 KK107238 EZA54792.1 SOQ45975.1 CVRI01000020 CRK91387.1 KK852498 KDR22678.1 GEZM01044876 JAV78072.1 KK854082 PTY10893.1 AJVK01005093 AJVK01005094 GFTR01006004 JAW10422.1 CCAG010017010 QOIP01000007 RLU21017.1 DS235371 EEB15388.1 PYGN01001874 PSN32376.1 GGMR01009451 MBY22070.1 ABLF02036147 GFXV01001361 MBW13166.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000037510
UP000053268
+ More
UP000095301 UP000076408 UP000215335 UP000095300 UP000037069 UP000075900 UP000002358 UP000053105 UP000078540 UP000008237 UP000005205 UP000075920 UP000075809 UP000075881 UP000075885 UP000000673 UP000075886 UP000075880 UP000075840 UP000092461 UP000030765 UP000008820 UP000075884 UP000007062 UP000069940 UP000249989 UP000076407 UP000069272 UP000091820 UP000078492 UP000242457 UP000002320 UP000005203 UP000235965 UP000078541 UP000078200 UP000092443 UP000092444 UP000009192 UP000007801 UP000192221 UP000002282 UP000007798 UP000000304 UP000092553 UP000008711 UP000000803 UP000001819 UP000001292 UP000008744 UP000268350 UP000001070 UP000008792 UP000007266 UP000076502 UP000075882 UP000030742 UP000019118 UP000053097 UP000192223 UP000183832 UP000027135 UP000092462 UP000279307 UP000009046 UP000245037 UP000007819
UP000095301 UP000076408 UP000215335 UP000095300 UP000037069 UP000075900 UP000002358 UP000053105 UP000078540 UP000008237 UP000005205 UP000075920 UP000075809 UP000075881 UP000075885 UP000000673 UP000075886 UP000075880 UP000075840 UP000092461 UP000030765 UP000008820 UP000075884 UP000007062 UP000069940 UP000249989 UP000076407 UP000069272 UP000091820 UP000078492 UP000242457 UP000002320 UP000005203 UP000235965 UP000078541 UP000078200 UP000092443 UP000092444 UP000009192 UP000007801 UP000192221 UP000002282 UP000007798 UP000000304 UP000092553 UP000008711 UP000000803 UP000001819 UP000001292 UP000008744 UP000268350 UP000001070 UP000008792 UP000007266 UP000076502 UP000075882 UP000030742 UP000019118 UP000053097 UP000192223 UP000183832 UP000027135 UP000092462 UP000279307 UP000009046 UP000245037 UP000007819
Interpro
IPR027750
TTLL1
+ More
IPR013815 ATP_grasp_subdomain_1
IPR004344 TTL/TTLL_fam
IPR036427 Bromodomain-like_sf
IPR018359 Bromodomain_CS
IPR017956 AT_hook_DNA-bd_motif
IPR001487 Bromodomain
IPR006212 Furin_repeat
IPR036642 Cyt_bc1_su8_sf
IPR004205 Cyt_bc1_su8
IPR013025 Ribosomal_L25/23
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR012678 Ribosomal_L23/L15e_core_dom_sf
IPR013815 ATP_grasp_subdomain_1
IPR004344 TTL/TTLL_fam
IPR036427 Bromodomain-like_sf
IPR018359 Bromodomain_CS
IPR017956 AT_hook_DNA-bd_motif
IPR001487 Bromodomain
IPR006212 Furin_repeat
IPR036642 Cyt_bc1_su8_sf
IPR004205 Cyt_bc1_su8
IPR013025 Ribosomal_L25/23
IPR012677 Nucleotide-bd_a/b_plait_sf
IPR012678 Ribosomal_L23/L15e_core_dom_sf
Gene 3D
CDD
ProteinModelPortal
H9JC13
A0A2A4K671
A0A2H1VZ45
A0A212FK33
A0A0N1IA08
A0A0L7KNZ4
+ More
A0A194QBN7 A0A0N1PIY6 H9JC14 A0A2A4K6G2 A0A194QCX3 A0A0L7L178 A0A2H1VYQ9 A0A1I8NH69 A0A336M6P3 A0A336KXR1 A0A182YE16 A0A232ETA9 A0A1I8PJ47 A0A0L0C9N8 A0A182R930 K7IYY3 A0A0M8ZWN3 A0A151I174 E2B7T0 A0A158NKC0 A0A0A1XII5 A0A182W857 W8C190 A0A0K8WGN2 A0A151WS30 A0A034WF49 A0A182JV02 A0A182P7N6 W5JMM9 A0A182QW01 A0A182IM81 A0A1S4H3P2 A0A182HIW0 A0A1B0GHE9 A0A084VJW0 Q17AY7 A0A182NQ30 Q7QDP1 A0A182GLP6 A0A3F2Z218 A0A182FZS6 A0A1A9W342 A0A195EIM7 A0A2A3E6H8 B0WZV2 A0A087ZY45 A0A310SCC6 A0A2J7PMN2 A0A195FBA5 A0A1A9VBY4 A0A1A9YDG0 A0A1B0G1Z0 B4KXA5 B3M9J3 A0A1W4VNM4 B4PIA7 B4MMY1 B4QR13 A0A0M3QVV4 B3NG56 Q9VZ97 Q29EA9 B4HU95 A0A1B6K0G9 B4H1S0 A0A1B6EK26 A0A3B0J736 B4IZ74 A0A1A9WDU0 B4MFZ2 D2A295 A0A2A4K5S3 A0A0L7KP30 A0A154P731 A0A182LGU7 U4U8A8 N6TVJ1 A0A026WFF6 A0A2H1VYQ7 A0A1W4XH13 A0A1J1HTR7 A0A067RI56 A0A1Y1LX05 A0A2R7VTP4 A0A1B0DCH7 A0A224XPD9 A0A1B0FAE4 A0A3L8DKS2 E0VPT2 A0A2P8XK43 H9JC09 A0A2S2NY11 J9JN31 A0A2H8TGB9
A0A194QBN7 A0A0N1PIY6 H9JC14 A0A2A4K6G2 A0A194QCX3 A0A0L7L178 A0A2H1VYQ9 A0A1I8NH69 A0A336M6P3 A0A336KXR1 A0A182YE16 A0A232ETA9 A0A1I8PJ47 A0A0L0C9N8 A0A182R930 K7IYY3 A0A0M8ZWN3 A0A151I174 E2B7T0 A0A158NKC0 A0A0A1XII5 A0A182W857 W8C190 A0A0K8WGN2 A0A151WS30 A0A034WF49 A0A182JV02 A0A182P7N6 W5JMM9 A0A182QW01 A0A182IM81 A0A1S4H3P2 A0A182HIW0 A0A1B0GHE9 A0A084VJW0 Q17AY7 A0A182NQ30 Q7QDP1 A0A182GLP6 A0A3F2Z218 A0A182FZS6 A0A1A9W342 A0A195EIM7 A0A2A3E6H8 B0WZV2 A0A087ZY45 A0A310SCC6 A0A2J7PMN2 A0A195FBA5 A0A1A9VBY4 A0A1A9YDG0 A0A1B0G1Z0 B4KXA5 B3M9J3 A0A1W4VNM4 B4PIA7 B4MMY1 B4QR13 A0A0M3QVV4 B3NG56 Q9VZ97 Q29EA9 B4HU95 A0A1B6K0G9 B4H1S0 A0A1B6EK26 A0A3B0J736 B4IZ74 A0A1A9WDU0 B4MFZ2 D2A295 A0A2A4K5S3 A0A0L7KP30 A0A154P731 A0A182LGU7 U4U8A8 N6TVJ1 A0A026WFF6 A0A2H1VYQ7 A0A1W4XH13 A0A1J1HTR7 A0A067RI56 A0A1Y1LX05 A0A2R7VTP4 A0A1B0DCH7 A0A224XPD9 A0A1B0FAE4 A0A3L8DKS2 E0VPT2 A0A2P8XK43 H9JC09 A0A2S2NY11 J9JN31 A0A2H8TGB9
PDB
4YLS
E-value=1.75079e-34,
Score=365
Ontologies
GO
PANTHER
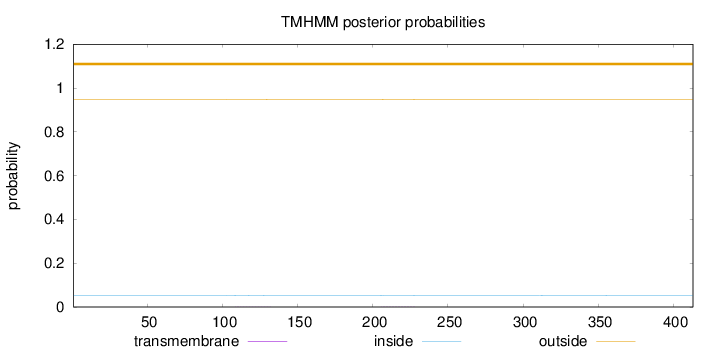
Topology
Length:
413
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02302
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05222
outside
1 - 413
Population Genetic Test Statistics
Pi
17.018707
Theta
62.234158
Tajima's D
-2.459191
CLR
479.110804
CSRT
0.000899955002249887
Interpretation
Uncertain
