Gene
KWMTBOMO10359
Annotation
PREDICTED:_alpha-(1?3)-fucosyltransferase_7-like_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.814
Sequence
CDS
ATGGAAAAAGTGAGTTACAGCCTTCAGTTGCACAAATATATTATCGATCAGTACTACGATTATCGAAAGGATCTGAAAACATCTAACCTAGGATCCATTCTCTTCAAAAGGAATGACAAGAATACATTATTAGTTGAAAAAAATAGAAAATTTACGGTACTAATATGGAGACATTGGGAATGGCTGAAATATCGCCACATTTTTCGCTATAGTTCAACACCGCATAATCCACTGGAGGATTGTAGTGTTAATAATTGTGAATTCACCGGAAACAACCGACTGTTAGACAAGGTCGACGCTGTGGTTGTTCATCTGCAAAAAGGAGTTCTCCCGAACATCTATAATAGAACAAGAACGCAGCGCTGGATATTTTTAACCGACGAATCTCCAATACATCTCTTTGGAAATGAAAAAAACAGACCAAACTTATCGGACTTGGTTAATGTCTTCAATTGGTCTATGACCTATAGAACTGATGCCGACATTCCTGTTCCATATGGTCGAACAATACCTCTTGAAGCTCCAATATTTACCGAAACAACAAATACGAATATTACTAAACTAATACCTAATTGGAAAAATAAAAGGCGAGATGTGTTGGCCTCGATTCTGATGTCTAACTGCGGTGTACCGTATCGAATGAAGTATTTAAATAAGTTGAAAAAACACATGAAAATTGATATTCATGGAAAATGCTCGGATTCACATCTTGTTAACAGCTGCCCTGGTCATTTCCGAGCAGATTGCAAGATAATATCAAACTACCTCTTTTACTTCGTATTAGAAAACTCCCTTTGTAAGGAATATATAACCGAGAAGTTGTTTCACCAAGCGTATGCAAAAGGGGCCATACCAATAATCCAAGGATCTGTGAAGCACTGA
Protein
MEKVSYSLQLHKYIIDQYYDYRKDLKTSNLGSILFKRNDKNTLLVEKNRKFTVLIWRHWEWLKYRHIFRYSSTPHNPLEDCSVNNCEFTGNNRLLDKVDAVVVHLQKGVLPNIYNRTRTQRWIFLTDESPIHLFGNEKNRPNLSDLVNVFNWSMTYRTDADIPVPYGRTIPLEAPIFTETTNTNITKLIPNWKNKRRDVLASILMSNCGVPYRMKYLNKLKKHMKIDIHGKCSDSHLVNSCPGHFRADCKIISNYLFYFVLENSLCKEYITEKLFHQAYAKGAIPIIQGSVKH
Summary
Similarity
Belongs to the glycosyltransferase 10 family.
Uniprot
A0A194QRI9
A0A194QBI8
A0A212FP91
A0A2A4JCX4
A0A2H1W952
A0A0L7KW49
+ More
J3JX77 N6SU16 A0A087THV8 V5IAT7 A0A2P8YPY1 A0A2L2Y7I9 A0A0K2UG84 A0A2L2Y7L4 A0A154PPW8 A0A2P6KIK7 A0A2J7REX0 Q05GU2 A0A088A5M2 D6WW27 F4WTL6 A0A0J7L2T3 A0A2P2I3P5 E9I934 A0A1Y1MSZ2 A0A195F1A8 A0A195CJU7 A0A067R6X2 A0A1V9XMQ8 E2AS11 A0A026X4U0 A0A195E830 A0A2A3E458 A0A0T6BGA4 A0A151I164 A0A151XB72 A0A158NG28 K7J5N2 A0A232FK45 A0A3R7PFF0 A0A0P4WHL7 A0A131XB30 A0A023GFR3 V5GGJ1 A0A1Z5LEX2 B7PIS7 A0A2J7REX3 A0A3R7NYV3 A0A2R5LCI7 A0A224YQP8 V5IHR6 L7MDR6 A0A131YYF0 A0A224YZ40 A0A1E1XR75 E2BJS3 G3MJ21 A0A1Y1MR45 L7MJF5 A0A310STK8 A0A0L7R2Y2 A0A0M9A7K4 A0A0P5BH63 A0A0P5WEJ8 A0A0P4YIR7 A0A0P5D7R1 A0A0P5BKJ6 A0A0P4YRD8 A0A0N8D3P2 E9H6Q7 A0A0V1JH26 A0A0V1KF12 A0A0N8BFB6 G3MZR2 A0A0P5QD24 A0A0P5ITZ0 L8J1X2 A0A0P5L6D4 A0A0P5RIE2 A0A0V1FB44 A0A0V1EYL6 A0A0P6FPQ1 A0A0P6EGV9 A0A0P5RL37 A0A0P5Q6W4 A0A0P6HDR3 A0A0P6EBR9 A0A0P5ZEQ7 A0A0P4Z1R1 A0A164ZU34 A0A0P5EGX4 A0A0P6BBL8 D2I3Y4 G1M4A1 A1XCQ0 A0A0P4Y6V7 A0A341CU57 F6YV26
J3JX77 N6SU16 A0A087THV8 V5IAT7 A0A2P8YPY1 A0A2L2Y7I9 A0A0K2UG84 A0A2L2Y7L4 A0A154PPW8 A0A2P6KIK7 A0A2J7REX0 Q05GU2 A0A088A5M2 D6WW27 F4WTL6 A0A0J7L2T3 A0A2P2I3P5 E9I934 A0A1Y1MSZ2 A0A195F1A8 A0A195CJU7 A0A067R6X2 A0A1V9XMQ8 E2AS11 A0A026X4U0 A0A195E830 A0A2A3E458 A0A0T6BGA4 A0A151I164 A0A151XB72 A0A158NG28 K7J5N2 A0A232FK45 A0A3R7PFF0 A0A0P4WHL7 A0A131XB30 A0A023GFR3 V5GGJ1 A0A1Z5LEX2 B7PIS7 A0A2J7REX3 A0A3R7NYV3 A0A2R5LCI7 A0A224YQP8 V5IHR6 L7MDR6 A0A131YYF0 A0A224YZ40 A0A1E1XR75 E2BJS3 G3MJ21 A0A1Y1MR45 L7MJF5 A0A310STK8 A0A0L7R2Y2 A0A0M9A7K4 A0A0P5BH63 A0A0P5WEJ8 A0A0P4YIR7 A0A0P5D7R1 A0A0P5BKJ6 A0A0P4YRD8 A0A0N8D3P2 E9H6Q7 A0A0V1JH26 A0A0V1KF12 A0A0N8BFB6 G3MZR2 A0A0P5QD24 A0A0P5ITZ0 L8J1X2 A0A0P5L6D4 A0A0P5RIE2 A0A0V1FB44 A0A0V1EYL6 A0A0P6FPQ1 A0A0P6EGV9 A0A0P5RL37 A0A0P5Q6W4 A0A0P6HDR3 A0A0P6EBR9 A0A0P5ZEQ7 A0A0P4Z1R1 A0A164ZU34 A0A0P5EGX4 A0A0P6BBL8 D2I3Y4 G1M4A1 A1XCQ0 A0A0P4Y6V7 A0A341CU57 F6YV26
Pubmed
26354079
22118469
26227816
22516182
23537049
29403074
+ More
26561354 17029591 18362917 19820115 21719571 21282665 28004739 24845553 28327890 20798317 24508170 30249741 21347285 20075255 28648823 28049606 25765539 28528879 28797301 25576852 26830274 29209593 22216098 21292972 19393038 22751099 20010809 12481130 15114417
26561354 17029591 18362917 19820115 21719571 21282665 28004739 24845553 28327890 20798317 24508170 30249741 21347285 20075255 28648823 28049606 25765539 28528879 28797301 25576852 26830274 29209593 22216098 21292972 19393038 22751099 20010809 12481130 15114417
EMBL
KQ461155
KPJ08107.1
KQ459232
KPJ02827.1
AGBW02003285
OWR55566.1
+ More
NWSH01001913 PCG69699.1 ODYU01007120 SOQ49611.1 JTDY01005122 KOB67350.1 BT127845 AEE62807.1 APGK01056896 APGK01056897 KB741277 KB632155 ENN71184.1 ERL89205.1 KK115300 KFM64697.1 GALX01000615 JAB67851.1 PYGN01000441 PSN46315.1 IAAA01012304 LAA04142.1 HACA01019365 CDW36726.1 IAAA01012303 LAA04138.1 KQ435022 KZC13922.1 MWRG01011080 PRD26134.1 NEVH01004429 PNF39384.1 AM279412 CAK50257.1 KQ971361 EFA08198.2 GL888341 EGI62437.1 LBMM01001190 KMQ96749.1 IACF01003007 LAB68639.1 GL761674 EFZ22915.1 GEZM01028131 GEZM01028129 GEZM01028128 JAV86447.1 KQ981864 KYN34166.1 KQ977721 KYN00344.1 KK852663 KDR19067.1 MNPL01007651 OQR74638.1 GL442209 EFN63785.1 KK107020 QOIP01000004 EZA62444.1 RLU23807.1 KQ979568 KYN20982.1 KZ288400 PBC26284.1 LJIG01000567 KRT86373.1 KQ976580 KYM79928.1 KQ982335 KYQ57633.1 ADTU01014941 ADTU01014942 NNAY01000089 OXU31094.1 QCYY01002503 ROT69896.1 GDRN01050265 JAI66475.1 GEFH01005223 JAP63358.1 GBBM01003833 JAC31585.1 GANP01015128 JAB69340.1 GFJQ02001017 JAW05953.1 ABJB010142677 DS721377 EEC06499.1 PNF39383.1 QCYY01002401 ROT70658.1 GGLE01003053 MBY07179.1 GFPF01008099 MAA19245.1 GANP01004472 JAB79996.1 GACK01003690 JAA61344.1 GEDV01004530 JAP84027.1 GFPF01008098 MAA19244.1 GFAA01001617 JAU01818.1 GL448636 EFN84048.1 JO841872 AEO33489.1 GEZM01028130 JAV86446.1 GACK01001715 JAA63319.1 KQ760116 OAD61948.1 KQ414665 KOC65136.1 KQ435732 KOX77553.1 GDIP01189992 JAJ33410.1 GDIP01087956 JAM15759.1 GDIP01228135 JAI95266.1 GDIP01161850 JAJ61552.1 GDIP01187438 JAJ35964.1 GDIP01223670 JAI99731.1 GDIP01071911 JAM31804.1 GL732598 EFX72552.1 JYDS01000004 KRZ34255.1 JYDV01000002 KRZ45794.1 GDIQ01183331 JAK68394.1 GDIQ01137186 JAL14540.1 GDIQ01233708 JAK18017.1 JH880295 ELR62711.1 GDIQ01183551 JAK68174.1 GDIQ01100344 JAL51382.1 JYDT01000144 KRY83321.1 JYDR01000002 KRY79066.1 GDIQ01076887 GDIQ01045117 JAN49620.1 GDIQ01069521 JAN25216.1 GDIQ01100861 JAL50865.1 GDIQ01118182 JAL33544.1 GDIQ01020670 JAN74067.1 GDIQ01065067 JAN29670.1 GDIP01045721 JAM57994.1 GDIP01231374 JAI92027.1 LRGB01000687 KZS16765.1 GDIP01146628 JAJ76774.1 GDIP01018813 JAM84902.1 GL194396 EFB23717.1 ACTA01076643 DQ339142 ABC68611.1 GDIP01232719 JAI90682.1
NWSH01001913 PCG69699.1 ODYU01007120 SOQ49611.1 JTDY01005122 KOB67350.1 BT127845 AEE62807.1 APGK01056896 APGK01056897 KB741277 KB632155 ENN71184.1 ERL89205.1 KK115300 KFM64697.1 GALX01000615 JAB67851.1 PYGN01000441 PSN46315.1 IAAA01012304 LAA04142.1 HACA01019365 CDW36726.1 IAAA01012303 LAA04138.1 KQ435022 KZC13922.1 MWRG01011080 PRD26134.1 NEVH01004429 PNF39384.1 AM279412 CAK50257.1 KQ971361 EFA08198.2 GL888341 EGI62437.1 LBMM01001190 KMQ96749.1 IACF01003007 LAB68639.1 GL761674 EFZ22915.1 GEZM01028131 GEZM01028129 GEZM01028128 JAV86447.1 KQ981864 KYN34166.1 KQ977721 KYN00344.1 KK852663 KDR19067.1 MNPL01007651 OQR74638.1 GL442209 EFN63785.1 KK107020 QOIP01000004 EZA62444.1 RLU23807.1 KQ979568 KYN20982.1 KZ288400 PBC26284.1 LJIG01000567 KRT86373.1 KQ976580 KYM79928.1 KQ982335 KYQ57633.1 ADTU01014941 ADTU01014942 NNAY01000089 OXU31094.1 QCYY01002503 ROT69896.1 GDRN01050265 JAI66475.1 GEFH01005223 JAP63358.1 GBBM01003833 JAC31585.1 GANP01015128 JAB69340.1 GFJQ02001017 JAW05953.1 ABJB010142677 DS721377 EEC06499.1 PNF39383.1 QCYY01002401 ROT70658.1 GGLE01003053 MBY07179.1 GFPF01008099 MAA19245.1 GANP01004472 JAB79996.1 GACK01003690 JAA61344.1 GEDV01004530 JAP84027.1 GFPF01008098 MAA19244.1 GFAA01001617 JAU01818.1 GL448636 EFN84048.1 JO841872 AEO33489.1 GEZM01028130 JAV86446.1 GACK01001715 JAA63319.1 KQ760116 OAD61948.1 KQ414665 KOC65136.1 KQ435732 KOX77553.1 GDIP01189992 JAJ33410.1 GDIP01087956 JAM15759.1 GDIP01228135 JAI95266.1 GDIP01161850 JAJ61552.1 GDIP01187438 JAJ35964.1 GDIP01223670 JAI99731.1 GDIP01071911 JAM31804.1 GL732598 EFX72552.1 JYDS01000004 KRZ34255.1 JYDV01000002 KRZ45794.1 GDIQ01183331 JAK68394.1 GDIQ01137186 JAL14540.1 GDIQ01233708 JAK18017.1 JH880295 ELR62711.1 GDIQ01183551 JAK68174.1 GDIQ01100344 JAL51382.1 JYDT01000144 KRY83321.1 JYDR01000002 KRY79066.1 GDIQ01076887 GDIQ01045117 JAN49620.1 GDIQ01069521 JAN25216.1 GDIQ01100861 JAL50865.1 GDIQ01118182 JAL33544.1 GDIQ01020670 JAN74067.1 GDIQ01065067 JAN29670.1 GDIP01045721 JAM57994.1 GDIP01231374 JAI92027.1 LRGB01000687 KZS16765.1 GDIP01146628 JAJ76774.1 GDIP01018813 JAM84902.1 GL194396 EFB23717.1 ACTA01076643 DQ339142 ABC68611.1 GDIP01232719 JAI90682.1
Proteomes
UP000053240
UP000053268
UP000007151
UP000218220
UP000037510
UP000019118
+ More
UP000030742 UP000054359 UP000245037 UP000076502 UP000235965 UP000005203 UP000007266 UP000007755 UP000036403 UP000078541 UP000078542 UP000027135 UP000192247 UP000000311 UP000053097 UP000279307 UP000078492 UP000242457 UP000078540 UP000075809 UP000005205 UP000002358 UP000215335 UP000283509 UP000001555 UP000008237 UP000053825 UP000053105 UP000000305 UP000054805 UP000054826 UP000009136 UP000054995 UP000054632 UP000076858 UP000008912 UP000252040 UP000008144
UP000030742 UP000054359 UP000245037 UP000076502 UP000235965 UP000005203 UP000007266 UP000007755 UP000036403 UP000078541 UP000078542 UP000027135 UP000192247 UP000000311 UP000053097 UP000279307 UP000078492 UP000242457 UP000078540 UP000075809 UP000005205 UP000002358 UP000215335 UP000283509 UP000001555 UP000008237 UP000053825 UP000053105 UP000000305 UP000054805 UP000054826 UP000009136 UP000054995 UP000054632 UP000076858 UP000008912 UP000252040 UP000008144
Interpro
Gene 3D
ProteinModelPortal
A0A194QRI9
A0A194QBI8
A0A212FP91
A0A2A4JCX4
A0A2H1W952
A0A0L7KW49
+ More
J3JX77 N6SU16 A0A087THV8 V5IAT7 A0A2P8YPY1 A0A2L2Y7I9 A0A0K2UG84 A0A2L2Y7L4 A0A154PPW8 A0A2P6KIK7 A0A2J7REX0 Q05GU2 A0A088A5M2 D6WW27 F4WTL6 A0A0J7L2T3 A0A2P2I3P5 E9I934 A0A1Y1MSZ2 A0A195F1A8 A0A195CJU7 A0A067R6X2 A0A1V9XMQ8 E2AS11 A0A026X4U0 A0A195E830 A0A2A3E458 A0A0T6BGA4 A0A151I164 A0A151XB72 A0A158NG28 K7J5N2 A0A232FK45 A0A3R7PFF0 A0A0P4WHL7 A0A131XB30 A0A023GFR3 V5GGJ1 A0A1Z5LEX2 B7PIS7 A0A2J7REX3 A0A3R7NYV3 A0A2R5LCI7 A0A224YQP8 V5IHR6 L7MDR6 A0A131YYF0 A0A224YZ40 A0A1E1XR75 E2BJS3 G3MJ21 A0A1Y1MR45 L7MJF5 A0A310STK8 A0A0L7R2Y2 A0A0M9A7K4 A0A0P5BH63 A0A0P5WEJ8 A0A0P4YIR7 A0A0P5D7R1 A0A0P5BKJ6 A0A0P4YRD8 A0A0N8D3P2 E9H6Q7 A0A0V1JH26 A0A0V1KF12 A0A0N8BFB6 G3MZR2 A0A0P5QD24 A0A0P5ITZ0 L8J1X2 A0A0P5L6D4 A0A0P5RIE2 A0A0V1FB44 A0A0V1EYL6 A0A0P6FPQ1 A0A0P6EGV9 A0A0P5RL37 A0A0P5Q6W4 A0A0P6HDR3 A0A0P6EBR9 A0A0P5ZEQ7 A0A0P4Z1R1 A0A164ZU34 A0A0P5EGX4 A0A0P6BBL8 D2I3Y4 G1M4A1 A1XCQ0 A0A0P4Y6V7 A0A341CU57 F6YV26
J3JX77 N6SU16 A0A087THV8 V5IAT7 A0A2P8YPY1 A0A2L2Y7I9 A0A0K2UG84 A0A2L2Y7L4 A0A154PPW8 A0A2P6KIK7 A0A2J7REX0 Q05GU2 A0A088A5M2 D6WW27 F4WTL6 A0A0J7L2T3 A0A2P2I3P5 E9I934 A0A1Y1MSZ2 A0A195F1A8 A0A195CJU7 A0A067R6X2 A0A1V9XMQ8 E2AS11 A0A026X4U0 A0A195E830 A0A2A3E458 A0A0T6BGA4 A0A151I164 A0A151XB72 A0A158NG28 K7J5N2 A0A232FK45 A0A3R7PFF0 A0A0P4WHL7 A0A131XB30 A0A023GFR3 V5GGJ1 A0A1Z5LEX2 B7PIS7 A0A2J7REX3 A0A3R7NYV3 A0A2R5LCI7 A0A224YQP8 V5IHR6 L7MDR6 A0A131YYF0 A0A224YZ40 A0A1E1XR75 E2BJS3 G3MJ21 A0A1Y1MR45 L7MJF5 A0A310STK8 A0A0L7R2Y2 A0A0M9A7K4 A0A0P5BH63 A0A0P5WEJ8 A0A0P4YIR7 A0A0P5D7R1 A0A0P5BKJ6 A0A0P4YRD8 A0A0N8D3P2 E9H6Q7 A0A0V1JH26 A0A0V1KF12 A0A0N8BFB6 G3MZR2 A0A0P5QD24 A0A0P5ITZ0 L8J1X2 A0A0P5L6D4 A0A0P5RIE2 A0A0V1FB44 A0A0V1EYL6 A0A0P6FPQ1 A0A0P6EGV9 A0A0P5RL37 A0A0P5Q6W4 A0A0P6HDR3 A0A0P6EBR9 A0A0P5ZEQ7 A0A0P4Z1R1 A0A164ZU34 A0A0P5EGX4 A0A0P6BBL8 D2I3Y4 G1M4A1 A1XCQ0 A0A0P4Y6V7 A0A341CU57 F6YV26
Ontologies
GO
PANTHER
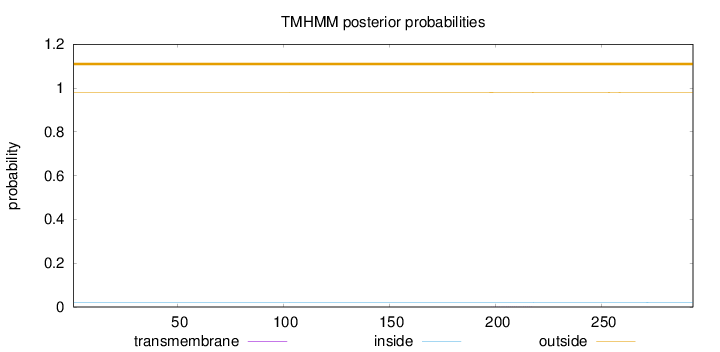
Topology
Subcellular location
Length:
293
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02193
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01943
outside
1 - 293
Population Genetic Test Statistics
Pi
175.758146
Theta
3.157186
Tajima's D
-1.800492
CLR
14.974217
CSRT
0.0218489075546223
Interpretation
Possibly Positive selection
