Gene
KWMTBOMO10358 Validated by peptides from experiments
Annotation
PREDICTED:_uncharacterized_protein_LOC106142516_isoform_X2_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 1.561 PlasmaMembrane Reliability : 1.412
Sequence
CDS
ATGGATGAACACACCTCAACGGACACAGTGCAAGAAAGCTCAAATCCAACATTACATGTTGAAGCACCAACAGATTGTGACTTTAACTTAACCCCTGTACAAATACAACAGCAATATTTTGAAAACTTAGGGGATTGTAACTGGATTAGATATGAAAAGTGCAATAGAAATCATTTATTGCAAAACTTGGCATTTGGACTATTTGGTCCTCCGATTACTGAAGGGGAGCTGATAGAAAAGGAAGAAATGAATGCAATTAATCTTCAAGGATACACTTCCAGACAAAAAGAAAAAGTTGACTTAGTCTATGCAAAAATATGTGAACAAGGTAAATATTCATTTGAAGATGAAATTTATATGTCGAATTTACTGGTTGTTTGTGCACGTCCAAAGCCCAAGAATTTTTTAAACATTCAACCGTCGAAATGTTGGATTGATCTACACGAACGGAGTGACTCCGAAATATGGCTCGTACCTGTATTTTGCATAAGAAAATGTATACCTATAGAAATGAATGCGAAACCATGCAAAATATATATTGATGACAATGCCCGGGTGTACCATAGTTGGAATGCTTACTTGACAGAAAACACATTGGACAAATGTGTTCTAATAGTCCCTGAGAACGGAGAATATACCGGAACACTTACTGAGGACGGAACAGACTTCACGGTGAATCTAACAGTGGCGCCGTCACCTGCCCTCGGACTGAAGTCACGCGTGCTGAGTACGGTGGACACAGCGACCACGGTGGTCTCGCTGAGCGCCGTGGGAGTGGTCGGGGTGGCCGCGCTAACACCGGTGGCGCCCGTGCTGCTCGCAAGCGCGGCCACCGTCTCCGTCACCGCGGGAATCTACGGGCTGATCAGGTCTTCGTTGCATTTACACGACCGAAGGACGCACGAACAGACCATAAACGTGACCAATTCTGAAGCTCGTAACAGTTGGTTAAACATAGCGGCCTCGGGCGTGGGTCTCACGGCCGGAGCGGCCAGCAATCTCCTGTCTAAATCAGCAGCGGCCGGCACCAACCTCACTACGTCGGGGAGGTTCTTGACGAAGTCAGTCGACGTGCTGCGACACGCGAACCCGTTGACCGGAGCTGCGAGCTTTACTAATGGCTTTGTGTATCTTATAATACAGTACAGGAAACACGGTCAGAAGCCGAGTCGTATTGAGATATTCCAATTCGCCATGTCGACGTTGTTCTTCTTCAACGCTGTCGTATCGAATCAGACGGCGCAAGAGATCATTGAGCAGAGTCAGGCGAACAAGATAAATGAGTTCCGCGATACGCTCCGGAGTAATCGGCACAGGAAAATTTTCGATAAGCTTTCCGCCGAAGCTAGACGAGTGCGGGGCACCGTTCAAGGTAACACCGAAGTCATCCAGGGCATCAAGAATATCGCTAACAAAGACGAATACTTTGCGCAAGTTCTGAGCATCAACAAGGACGTGAACCAGCACAGGCTCAGGATATCGATGACAGCCGACGGCCGGGTCATGTTGAACAACCAACACGGGTACAACCCGTCCGAGTTACACTCGCTCGGCTCGGAAGGGAGATCGCAAATATTCACCGAAATCGGCCCAGCCCCCCCGGGGCCTGCTAATGTGTCCACACGTCTGAACTCGCCCGTCAACAGAGTGGACAACTACTTCGGAGTAGACGACGAAGAAGGCCATTTATTAGTCGGCATCCGTCCAGAAGAGATACTCCAAATCGGCACATATTTCGTGCGTATCAGCTCGTCGGGCACCGATGAAATAGTTTCGATGCTCCAAAATCTCTCCGAAGAGGTCTATACGAATTTGATGACCCTATCCTTTAATCTGCTGTCGAAACTTGTGCCAAGCGAAATTGCCAATCTGAAGTTGCTCAGTCCGGAGCGAGATTTGATTTCGCAGGTAGTTGAATTCGTTTTCAATTACATGAAACAGCGGAGACCGTTCGGGGAAGCGAGACATGACGATAACGGAATCGTTGAAGTAGTAAAAGACTTCTTTCACGATGGCCGAGTTAATCAAGACACGATATTGCGATTGAAGAAAGATTTAATCGAATGGCTGGACGATGAAATTACACGTAAAAATGAGCTAAATCCTAATAAGAAATCCTTAATCTGCAATGTGTGTAAAGGAGTTAGATACGCTTAA
Protein
MDEHTSTDTVQESSNPTLHVEAPTDCDFNLTPVQIQQQYFENLGDCNWIRYEKCNRNHLLQNLAFGLFGPPITEGELIEKEEMNAINLQGYTSRQKEKVDLVYAKICEQGKYSFEDEIYMSNLLVVCARPKPKNFLNIQPSKCWIDLHERSDSEIWLVPVFCIRKCIPIEMNAKPCKIYIDDNARVYHSWNAYLTENTLDKCVLIVPENGEYTGTLTEDGTDFTVNLTVAPSPALGLKSRVLSTVDTATTVVSLSAVGVVGVAALTPVAPVLLASAATVSVTAGIYGLIRSSLHLHDRRTHEQTINVTNSEARNSWLNIAASGVGLTAGAASNLLSKSAAAGTNLTTSGRFLTKSVDVLRHANPLTGAASFTNGFVYLIIQYRKHGQKPSRIEIFQFAMSTLFFFNAVVSNQTAQEIIEQSQANKINEFRDTLRSNRHRKIFDKLSAEARRVRGTVQGNTEVIQGIKNIANKDEYFAQVLSINKDVNQHRLRISMTADGRVMLNNQHGYNPSELHSLGSEGRSQIFTEIGPAPPGPANVSTRLNSPVNRVDNYFGVDDEEGHLLVGIRPEEILQIGTYFVRISSSGTDEIVSMLQNLSEEVYTNLMTLSFNLLSKLVPSEIANLKLLSPERDLISQVVEFVFNYMKQRRPFGEARHDDNGIVEVVKDFFHDGRVNQDTILRLKKDLIEWLDDEITRKNELNPNKKSLICNVCKGVRYA
Summary
Uniprot
A0A2A4JDE4
A0A2H1WN45
A0A3S2M4L7
A0A194QHA7
A0A194QRH2
A0A0L7KNQ7
+ More
A0A2A4JC77 A0A2H1VTZ8 D6WTN8 A0A1W4XCY6 A0A1W4XBU6 D6WUF4 A0A1Y1LM37 A0A139WG29 A0A1W4WYD5 A0A0K8W5J4 A0A0K8UFM2 A0A1B6J932 K7JF86 A0A1B6J5V5 A0A232F1B8 W8BJT6 W8C5B0 W8CBB2 A0A0A1XHE4 A0A0J7NF99 A0A1I8Q4C6 A0A0M4EI97 Q2M145 B4N3X3 A0A1B6M3D4 B4GUP0 A0A0J9RQB5 B4QKD3 B3M6X3 E9IUV7
A0A2A4JC77 A0A2H1VTZ8 D6WTN8 A0A1W4XCY6 A0A1W4XBU6 D6WUF4 A0A1Y1LM37 A0A139WG29 A0A1W4WYD5 A0A0K8W5J4 A0A0K8UFM2 A0A1B6J932 K7JF86 A0A1B6J5V5 A0A232F1B8 W8BJT6 W8C5B0 W8CBB2 A0A0A1XHE4 A0A0J7NF99 A0A1I8Q4C6 A0A0M4EI97 Q2M145 B4N3X3 A0A1B6M3D4 B4GUP0 A0A0J9RQB5 B4QKD3 B3M6X3 E9IUV7
Pubmed
EMBL
NWSH01001913
PCG69698.1
ODYU01009816
SOQ54491.1
RSAL01000036
RVE51276.1
+ More
KQ459232 KPJ02826.1 KQ461155 KPJ08108.1 JTDY01007776 KOB64948.1 PCG69697.1 ODYU01004412 SOQ44258.1 KQ971355 EFA07204.2 KQ971352 EFA07295.2 GEZM01051772 JAV74709.1 KYB26757.1 GDHF01017990 GDHF01006000 JAI34324.1 JAI46314.1 GDHF01026938 GDHF01012558 GDHF01010695 JAI25376.1 JAI39756.1 JAI41619.1 GECU01012012 JAS95694.1 AAZX01007293 GECU01013172 GECU01000719 JAS94534.1 JAT06988.1 NNAY01001284 OXU24491.1 GAMC01004985 JAC01571.1 GAMC01004984 JAC01572.1 GAMC01004986 JAC01570.1 GBXI01003967 JAD10325.1 LBMM01005788 KMQ91205.1 CP012525 ALC43551.1 CH379069 EAL30731.2 CH964095 EDW79328.2 GEBQ01009554 JAT30423.1 CH479191 EDW26323.1 CM002912 KMY98071.1 CM000363 EDX09556.1 CH902618 EDV39809.1 GL766058 EFZ15639.1
KQ459232 KPJ02826.1 KQ461155 KPJ08108.1 JTDY01007776 KOB64948.1 PCG69697.1 ODYU01004412 SOQ44258.1 KQ971355 EFA07204.2 KQ971352 EFA07295.2 GEZM01051772 JAV74709.1 KYB26757.1 GDHF01017990 GDHF01006000 JAI34324.1 JAI46314.1 GDHF01026938 GDHF01012558 GDHF01010695 JAI25376.1 JAI39756.1 JAI41619.1 GECU01012012 JAS95694.1 AAZX01007293 GECU01013172 GECU01000719 JAS94534.1 JAT06988.1 NNAY01001284 OXU24491.1 GAMC01004985 JAC01571.1 GAMC01004984 JAC01572.1 GAMC01004986 JAC01570.1 GBXI01003967 JAD10325.1 LBMM01005788 KMQ91205.1 CP012525 ALC43551.1 CH379069 EAL30731.2 CH964095 EDW79328.2 GEBQ01009554 JAT30423.1 CH479191 EDW26323.1 CM002912 KMY98071.1 CM000363 EDX09556.1 CH902618 EDV39809.1 GL766058 EFZ15639.1
Proteomes
Pfam
Interpro
IPR031962
DUF4781
+ More
IPR035437 SNase_OB-fold_sf
IPR002999 Tudor
IPR029071 Ubiquitin-like_domsf
IPR006577 UAS
IPR027417 P-loop_NTPase
IPR020003 ATPase_a/bsu_AS
IPR022878 V-ATPase_asu
IPR000194 ATPase_F1/V1/A1_a/bsu_nucl-bd
IPR024034 ATPase_F1/V1_b/a_C
IPR036121 ATPase_F1/V1/A1_a/bsu_N_sf
IPR023366 ATP_synth_asu-like_sf
IPR031686 ATP-synth_a_Xtn
IPR005725 ATPase_V1-cplx_asu
IPR004100 ATPase_F1/V1/A1_a/bsu_N
IPR035437 SNase_OB-fold_sf
IPR002999 Tudor
IPR029071 Ubiquitin-like_domsf
IPR006577 UAS
IPR027417 P-loop_NTPase
IPR020003 ATPase_a/bsu_AS
IPR022878 V-ATPase_asu
IPR000194 ATPase_F1/V1/A1_a/bsu_nucl-bd
IPR024034 ATPase_F1/V1_b/a_C
IPR036121 ATPase_F1/V1/A1_a/bsu_N_sf
IPR023366 ATP_synth_asu-like_sf
IPR031686 ATP-synth_a_Xtn
IPR005725 ATPase_V1-cplx_asu
IPR004100 ATPase_F1/V1/A1_a/bsu_N
Gene 3D
ProteinModelPortal
A0A2A4JDE4
A0A2H1WN45
A0A3S2M4L7
A0A194QHA7
A0A194QRH2
A0A0L7KNQ7
+ More
A0A2A4JC77 A0A2H1VTZ8 D6WTN8 A0A1W4XCY6 A0A1W4XBU6 D6WUF4 A0A1Y1LM37 A0A139WG29 A0A1W4WYD5 A0A0K8W5J4 A0A0K8UFM2 A0A1B6J932 K7JF86 A0A1B6J5V5 A0A232F1B8 W8BJT6 W8C5B0 W8CBB2 A0A0A1XHE4 A0A0J7NF99 A0A1I8Q4C6 A0A0M4EI97 Q2M145 B4N3X3 A0A1B6M3D4 B4GUP0 A0A0J9RQB5 B4QKD3 B3M6X3 E9IUV7
A0A2A4JC77 A0A2H1VTZ8 D6WTN8 A0A1W4XCY6 A0A1W4XBU6 D6WUF4 A0A1Y1LM37 A0A139WG29 A0A1W4WYD5 A0A0K8W5J4 A0A0K8UFM2 A0A1B6J932 K7JF86 A0A1B6J5V5 A0A232F1B8 W8BJT6 W8C5B0 W8CBB2 A0A0A1XHE4 A0A0J7NF99 A0A1I8Q4C6 A0A0M4EI97 Q2M145 B4N3X3 A0A1B6M3D4 B4GUP0 A0A0J9RQB5 B4QKD3 B3M6X3 E9IUV7
Ontologies
GO
PANTHER
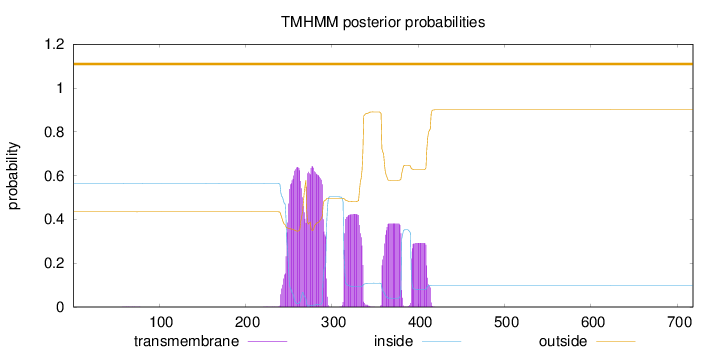
Topology
Length:
718
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
49.28567
Exp number, first 60 AAs:
0.00172
Total prob of N-in:
0.56534
outside
1 - 718
Population Genetic Test Statistics
Pi
40.915077
Theta
152.825306
Tajima's D
-0.955261
CLR
0.584111
CSRT
0.145642717864107
Interpretation
Uncertain
