Gene
KWMTBOMO10347
Annotation
PREDICTED:_phosphatidylinositol_N-acetylglucosaminyltransferase_subunit_Q_isoform_X2_[Papilio_polytes]
Full name
Phosphatidylinositol N-acetylglucosaminyltransferase subunit Q
Alternative Name
MGpi1p
N-acetylglucosamyl transferase component GPI1
Phosphatidylinositol-glycan biosynthesis class Q protein
N-acetylglucosamyl transferase component GPI1
Phosphatidylinositol-glycan biosynthesis class Q protein
Location in the cell
PlasmaMembrane Reliability : 4.846
Sequence
CDS
ATGTTAGGAAAATATTTTTCTTACCACGTTCAGCTGTGGTGGCTGTTTTTAGATGTTTCGGGAGAAAAACTTGACTTAATACTACAGGTCTACCGATATGTGGGTTATTTAGGGCTCACGTTTCAAATAGCAGTGATTTCGGATATGATATGCATTGCAACGTTTCATTCATATTGCATCTATGTTTATGCTGCAAGATTGCTTAACATGCAAATATCAGGTCTGATTGCATTGTTGAGACTCTTCGTTGGTCGCAAATACAATCCACTGAGGGGCAGCATAGACTCTTGTGAATATACGAATCAAGAATTATTTGTAGGAACCGTGGCTTTTACAATATTGTTGCTTTTGCTGCCCACAACAGCATTGTATTATATTGTGTTTACACTGTTTAGAATACTATCTCTGGCGCTTCAGTATATATTAGCGCAGCTGATCTACATTATACAAACATTCCCGTTTTATGTGATATCCCTCTGGGTGTTAAAATCGCCAAAAGTAGCAGGAAACGTTTTAATAGAAAGAGTGAATGGTAGAAACTCACCTTTAGTTGTGGAGGTGCAATTGTTGAGCAAATCCTTAACAGATTTGGTAGCCATGTTCAGACCGCCCGTTGAAAAACCGAAAAATCTTGAATGGACCAATCTATTATCCAATGTTCTAGTTGGAAAACAAATTGTATAA
Protein
MLGKYFSYHVQLWWLFLDVSGEKLDLILQVYRYVGYLGLTFQIAVISDMICIATFHSYCIYVYAARLLNMQISGLIALLRLFVGRKYNPLRGSIDSCEYTNQELFVGTVAFTILLLLLPTTALYYIVFTLFRILSLALQYILAQLIYIIQTFPFYVISLWVLKSPKVAGNVLIERVNGRNSPLVVEVQLLSKSLTDLVAMFRPPVEKPKNLEWTNLLSNVLVGKQIV
Summary
Catalytic Activity
a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol) + UDP-N-acetyl-alpha-D-glucosamine = a 6-(N-acetyl-alpha-D-glucosaminyl)-1-phosphatidyl-1D-myo-inositol + H(+) + UDP
Subunit
Associates with PIGA, PIGC, PIGH, PIGP and DPM2. The latter is not essential for activity.
Similarity
Belongs to the PIGQ family.
Keywords
Complete proteome
Glycosyltransferase
GPI-anchor biosynthesis
Membrane
Reference proteome
Transferase
Transmembrane
Transmembrane helix
Feature
chain Phosphatidylinositol N-acetylglucosaminyltransferase subunit Q
Uniprot
A0A1E1WQB3
A0A2A4JUZ3
A0A2H1W2Z0
S4P5J0
A0A067RQR3
A0A2P8ZKY8
+ More
A0A2J7QJF5 A0A139WCV9 A0A1B6DJS4 E9IEF2 A0A151ISH5 A0A3L8DUQ1 A0A026X0V2 F4X5R7 A0A158NMC4 A0A2A3EB01 K7JKT6 A0A232EQ93 A0A146LI72 A0A151WVG4 A0A195CCL3 A0A1Y1NFC8 A0A088AMI3 A0A1B6GIT5 E2BHH6 A0A1B6MFZ7 A0A151K1C4 A0A0T6BEY1 A0A0A9XU87 A0A0M9A2C0 A0A151I387 A0A1W4X273 A0A0L7REY8 A0A0P4VKJ1 J9M0C0 A0A154P1J6 A0A0P5HJ97 A0A0P5HN41 A0A0P5TN65 A0A0N8AH51 A0A0P5YWC7 A0A0N8ANI0 A0A0P5DHZ0 A0A0P5E8Y4 R4WDW8 A0A0P6BYU9 A0A1S3K4Q5 A0A0P5PN85 A0A0P5ELU0 A0A0P5LJA2 A0A094LBX6 A0A226NSJ1 T1KNI6 A0A2I4AK31 F7BRH8 A0A091T8R0 A0A384A3F4 A0A340X3E9 A0A341CLC0 A0A341CLA2 A0A384A3P4 A0A091IMH7 A0A091WMW3 S9XHP1 A0A093QS79 A0A091LB83 A0A091M3S6 A0A3Q0D9N3 A0A1S3EST9 A0A093F7G3 A0A091RR99 A0A087QI88 A0A093FK64 A0A2Y9Q3D8 A0A287D5M4 G3HBT3 A0A093LS95 A0A0A0AVC2 Q3TF56 Q9QYT7 A0A091URH0 G1SE00 A0A2Y9PX26 A0A2Y9PY12 A0A091UCQ0 A0A093NBB5 A0A1S3ESW2 A0A091PMS0 A0A286XXY3 S7N043 A0A226MW60 A0A2Y9DEM2 A0A2K6ENX2 A0A091Q3F7 E1BVN7 A0A384CKB0 Q642B8 A0A091NLB8
A0A2J7QJF5 A0A139WCV9 A0A1B6DJS4 E9IEF2 A0A151ISH5 A0A3L8DUQ1 A0A026X0V2 F4X5R7 A0A158NMC4 A0A2A3EB01 K7JKT6 A0A232EQ93 A0A146LI72 A0A151WVG4 A0A195CCL3 A0A1Y1NFC8 A0A088AMI3 A0A1B6GIT5 E2BHH6 A0A1B6MFZ7 A0A151K1C4 A0A0T6BEY1 A0A0A9XU87 A0A0M9A2C0 A0A151I387 A0A1W4X273 A0A0L7REY8 A0A0P4VKJ1 J9M0C0 A0A154P1J6 A0A0P5HJ97 A0A0P5HN41 A0A0P5TN65 A0A0N8AH51 A0A0P5YWC7 A0A0N8ANI0 A0A0P5DHZ0 A0A0P5E8Y4 R4WDW8 A0A0P6BYU9 A0A1S3K4Q5 A0A0P5PN85 A0A0P5ELU0 A0A0P5LJA2 A0A094LBX6 A0A226NSJ1 T1KNI6 A0A2I4AK31 F7BRH8 A0A091T8R0 A0A384A3F4 A0A340X3E9 A0A341CLC0 A0A341CLA2 A0A384A3P4 A0A091IMH7 A0A091WMW3 S9XHP1 A0A093QS79 A0A091LB83 A0A091M3S6 A0A3Q0D9N3 A0A1S3EST9 A0A093F7G3 A0A091RR99 A0A087QI88 A0A093FK64 A0A2Y9Q3D8 A0A287D5M4 G3HBT3 A0A093LS95 A0A0A0AVC2 Q3TF56 Q9QYT7 A0A091URH0 G1SE00 A0A2Y9PX26 A0A2Y9PY12 A0A091UCQ0 A0A093NBB5 A0A1S3ESW2 A0A091PMS0 A0A286XXY3 S7N043 A0A226MW60 A0A2Y9DEM2 A0A2K6ENX2 A0A091Q3F7 E1BVN7 A0A384CKB0 Q642B8 A0A091NLB8
EC Number
2.4.1.198
Pubmed
23622113
24845553
29403074
18362917
19820115
21282665
+ More
30249741 24508170 21719571 21347285 20075255 28648823 26823975 28004739 20798317 25401762 27129103 23691247 24621616 19892987 23149746 28071753 21804562 10349636 11042159 11076861 11217851 12466851 12040188 16141073 21183079 9729469 10373468 15489334 21993624 15592404 15057822 15632090
30249741 24508170 21719571 21347285 20075255 28648823 26823975 28004739 20798317 25401762 27129103 23691247 24621616 19892987 23149746 28071753 21804562 10349636 11042159 11076861 11217851 12466851 12040188 16141073 21183079 9729469 10373468 15489334 21993624 15592404 15057822 15632090
EMBL
GDQN01001890
JAT89164.1
NWSH01000569
PCG75586.1
ODYU01005919
SOQ47316.1
+ More
GAIX01005514 JAA87046.1 KK852478 KDR22985.1 PYGN01000026 PSN57164.1 NEVH01013555 PNF28725.1 KQ971363 KYB25715.1 GEDC01011367 JAS25931.1 GL762591 EFZ21036.1 KQ981090 KYN09564.1 QOIP01000004 RLU24006.1 KK107054 EZA61601.1 GL888742 EGI58157.1 ADTU01020384 KZ288313 PBC28366.1 NNAY01002812 OXU20524.1 GDHC01010785 JAQ07844.1 KQ982701 KYQ51900.1 KQ977957 KYM98455.1 GEZM01003860 JAV96521.1 GECZ01007439 JAS62330.1 GL448301 EFN84829.1 GEBQ01005145 JAT34832.1 KQ981175 KYN45294.1 LJIG01001044 KRT85896.1 GBHO01021148 GBHO01021146 GBRD01016970 GBRD01010658 JAG22456.1 JAG22458.1 JAG48857.1 KQ435791 KOX74389.1 KQ976507 KYM82787.1 KQ414608 KOC69379.1 GDKW01000996 JAI55599.1 ABLF02031485 KQ434796 KZC05727.1 GDIQ01226149 JAK25576.1 GDIQ01224996 JAK26729.1 GDIP01125298 JAL78416.1 GDIP01147929 JAJ75473.1 GDIP01065691 GDIQ01044952 LRGB01000687 JAM38024.1 JAN49785.1 KZS16823.1 GDIQ01258419 JAJ93305.1 GDIP01155931 JAJ67471.1 GDIP01146584 JAJ76818.1 AK417929 BAN21144.1 GDIP01012237 JAM91478.1 GDIQ01131689 JAL20037.1 GDIP01157893 JAJ65509.1 GDIQ01170967 JAK80758.1 KL284272 KFZ68275.1 AWGT02000910 OXB70886.1 CAEY01000277 KK440279 KFQ69997.1 KK500657 KFP09537.1 KK735903 KFR16455.1 KB016354 EPY90077.1 KL421925 KFW89205.1 KK745998 KFP40211.1 KK521056 KFP66433.1 KK384261 KFV50151.1 KK930700 KFQ44983.1 KL225618 KFM00942.1 KK629051 KFV54679.1 AGTP01030746 JH000273 EGW04370.1 KK610035 KFW11556.1 KL873367 KGL98504.1 AK163680 AK169284 CH466606 BAE37455.1 BAE41042.1 EDL22475.1 AF030178 AB008895 AB008921 BC014287 BC003917 KL410017 KFQ93281.1 KK428087 KFQ88296.1 KL224601 KFW61476.1 KK662202 KFQ08950.1 AAKN02007386 KE162966 EPQ10274.1 MCFN01000402 OXB59269.1 KK685658 KFQ14455.1 AADN05000376 AC126071 BC081875 CH473948 AAH81875.1 EDM03983.1 KL385218 KFP89684.1
GAIX01005514 JAA87046.1 KK852478 KDR22985.1 PYGN01000026 PSN57164.1 NEVH01013555 PNF28725.1 KQ971363 KYB25715.1 GEDC01011367 JAS25931.1 GL762591 EFZ21036.1 KQ981090 KYN09564.1 QOIP01000004 RLU24006.1 KK107054 EZA61601.1 GL888742 EGI58157.1 ADTU01020384 KZ288313 PBC28366.1 NNAY01002812 OXU20524.1 GDHC01010785 JAQ07844.1 KQ982701 KYQ51900.1 KQ977957 KYM98455.1 GEZM01003860 JAV96521.1 GECZ01007439 JAS62330.1 GL448301 EFN84829.1 GEBQ01005145 JAT34832.1 KQ981175 KYN45294.1 LJIG01001044 KRT85896.1 GBHO01021148 GBHO01021146 GBRD01016970 GBRD01010658 JAG22456.1 JAG22458.1 JAG48857.1 KQ435791 KOX74389.1 KQ976507 KYM82787.1 KQ414608 KOC69379.1 GDKW01000996 JAI55599.1 ABLF02031485 KQ434796 KZC05727.1 GDIQ01226149 JAK25576.1 GDIQ01224996 JAK26729.1 GDIP01125298 JAL78416.1 GDIP01147929 JAJ75473.1 GDIP01065691 GDIQ01044952 LRGB01000687 JAM38024.1 JAN49785.1 KZS16823.1 GDIQ01258419 JAJ93305.1 GDIP01155931 JAJ67471.1 GDIP01146584 JAJ76818.1 AK417929 BAN21144.1 GDIP01012237 JAM91478.1 GDIQ01131689 JAL20037.1 GDIP01157893 JAJ65509.1 GDIQ01170967 JAK80758.1 KL284272 KFZ68275.1 AWGT02000910 OXB70886.1 CAEY01000277 KK440279 KFQ69997.1 KK500657 KFP09537.1 KK735903 KFR16455.1 KB016354 EPY90077.1 KL421925 KFW89205.1 KK745998 KFP40211.1 KK521056 KFP66433.1 KK384261 KFV50151.1 KK930700 KFQ44983.1 KL225618 KFM00942.1 KK629051 KFV54679.1 AGTP01030746 JH000273 EGW04370.1 KK610035 KFW11556.1 KL873367 KGL98504.1 AK163680 AK169284 CH466606 BAE37455.1 BAE41042.1 EDL22475.1 AF030178 AB008895 AB008921 BC014287 BC003917 KL410017 KFQ93281.1 KK428087 KFQ88296.1 KL224601 KFW61476.1 KK662202 KFQ08950.1 AAKN02007386 KE162966 EPQ10274.1 MCFN01000402 OXB59269.1 KK685658 KFQ14455.1 AADN05000376 AC126071 BC081875 CH473948 AAH81875.1 EDM03983.1 KL385218 KFP89684.1
Proteomes
UP000218220
UP000027135
UP000245037
UP000235965
UP000007266
UP000078492
+ More
UP000279307 UP000053097 UP000007755 UP000005205 UP000242457 UP000002358 UP000215335 UP000075809 UP000078542 UP000005203 UP000008237 UP000078541 UP000053105 UP000078540 UP000192223 UP000053825 UP000007819 UP000076502 UP000076858 UP000085678 UP000198419 UP000015104 UP000192220 UP000002281 UP000261681 UP000265300 UP000252040 UP000053119 UP000053605 UP000189706 UP000081671 UP000053286 UP000248483 UP000005215 UP000001075 UP000053858 UP000000589 UP000053283 UP000001811 UP000054081 UP000005447 UP000198323 UP000248480 UP000233160 UP000000539 UP000261680 UP000002494
UP000279307 UP000053097 UP000007755 UP000005205 UP000242457 UP000002358 UP000215335 UP000075809 UP000078542 UP000005203 UP000008237 UP000078541 UP000053105 UP000078540 UP000192223 UP000053825 UP000007819 UP000076502 UP000076858 UP000085678 UP000198419 UP000015104 UP000192220 UP000002281 UP000261681 UP000265300 UP000252040 UP000053119 UP000053605 UP000189706 UP000081671 UP000053286 UP000248483 UP000005215 UP000001075 UP000053858 UP000000589 UP000053283 UP000001811 UP000054081 UP000005447 UP000198323 UP000248480 UP000233160 UP000000539 UP000261680 UP000002494
PRIDE
ProteinModelPortal
A0A1E1WQB3
A0A2A4JUZ3
A0A2H1W2Z0
S4P5J0
A0A067RQR3
A0A2P8ZKY8
+ More
A0A2J7QJF5 A0A139WCV9 A0A1B6DJS4 E9IEF2 A0A151ISH5 A0A3L8DUQ1 A0A026X0V2 F4X5R7 A0A158NMC4 A0A2A3EB01 K7JKT6 A0A232EQ93 A0A146LI72 A0A151WVG4 A0A195CCL3 A0A1Y1NFC8 A0A088AMI3 A0A1B6GIT5 E2BHH6 A0A1B6MFZ7 A0A151K1C4 A0A0T6BEY1 A0A0A9XU87 A0A0M9A2C0 A0A151I387 A0A1W4X273 A0A0L7REY8 A0A0P4VKJ1 J9M0C0 A0A154P1J6 A0A0P5HJ97 A0A0P5HN41 A0A0P5TN65 A0A0N8AH51 A0A0P5YWC7 A0A0N8ANI0 A0A0P5DHZ0 A0A0P5E8Y4 R4WDW8 A0A0P6BYU9 A0A1S3K4Q5 A0A0P5PN85 A0A0P5ELU0 A0A0P5LJA2 A0A094LBX6 A0A226NSJ1 T1KNI6 A0A2I4AK31 F7BRH8 A0A091T8R0 A0A384A3F4 A0A340X3E9 A0A341CLC0 A0A341CLA2 A0A384A3P4 A0A091IMH7 A0A091WMW3 S9XHP1 A0A093QS79 A0A091LB83 A0A091M3S6 A0A3Q0D9N3 A0A1S3EST9 A0A093F7G3 A0A091RR99 A0A087QI88 A0A093FK64 A0A2Y9Q3D8 A0A287D5M4 G3HBT3 A0A093LS95 A0A0A0AVC2 Q3TF56 Q9QYT7 A0A091URH0 G1SE00 A0A2Y9PX26 A0A2Y9PY12 A0A091UCQ0 A0A093NBB5 A0A1S3ESW2 A0A091PMS0 A0A286XXY3 S7N043 A0A226MW60 A0A2Y9DEM2 A0A2K6ENX2 A0A091Q3F7 E1BVN7 A0A384CKB0 Q642B8 A0A091NLB8
A0A2J7QJF5 A0A139WCV9 A0A1B6DJS4 E9IEF2 A0A151ISH5 A0A3L8DUQ1 A0A026X0V2 F4X5R7 A0A158NMC4 A0A2A3EB01 K7JKT6 A0A232EQ93 A0A146LI72 A0A151WVG4 A0A195CCL3 A0A1Y1NFC8 A0A088AMI3 A0A1B6GIT5 E2BHH6 A0A1B6MFZ7 A0A151K1C4 A0A0T6BEY1 A0A0A9XU87 A0A0M9A2C0 A0A151I387 A0A1W4X273 A0A0L7REY8 A0A0P4VKJ1 J9M0C0 A0A154P1J6 A0A0P5HJ97 A0A0P5HN41 A0A0P5TN65 A0A0N8AH51 A0A0P5YWC7 A0A0N8ANI0 A0A0P5DHZ0 A0A0P5E8Y4 R4WDW8 A0A0P6BYU9 A0A1S3K4Q5 A0A0P5PN85 A0A0P5ELU0 A0A0P5LJA2 A0A094LBX6 A0A226NSJ1 T1KNI6 A0A2I4AK31 F7BRH8 A0A091T8R0 A0A384A3F4 A0A340X3E9 A0A341CLC0 A0A341CLA2 A0A384A3P4 A0A091IMH7 A0A091WMW3 S9XHP1 A0A093QS79 A0A091LB83 A0A091M3S6 A0A3Q0D9N3 A0A1S3EST9 A0A093F7G3 A0A091RR99 A0A087QI88 A0A093FK64 A0A2Y9Q3D8 A0A287D5M4 G3HBT3 A0A093LS95 A0A0A0AVC2 Q3TF56 Q9QYT7 A0A091URH0 G1SE00 A0A2Y9PX26 A0A2Y9PY12 A0A091UCQ0 A0A093NBB5 A0A1S3ESW2 A0A091PMS0 A0A286XXY3 S7N043 A0A226MW60 A0A2Y9DEM2 A0A2K6ENX2 A0A091Q3F7 E1BVN7 A0A384CKB0 Q642B8 A0A091NLB8
Ontologies
PATHWAY
GO
PANTHER
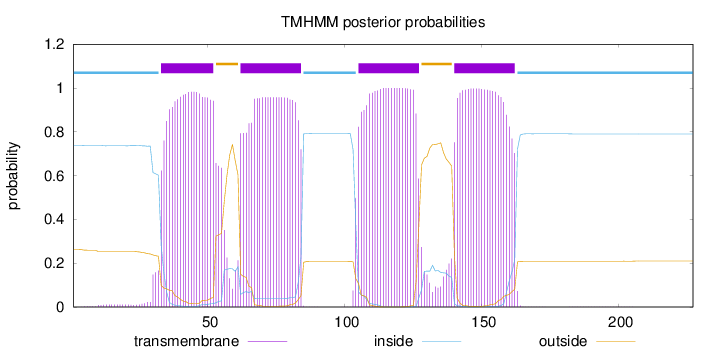
Topology
Subcellular location
Membrane
Length:
227
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
89.2052600000001
Exp number, first 60 AAs:
22.12574
Total prob of N-in:
0.73814
POSSIBLE N-term signal
sequence
inside
1 - 32
TMhelix
33 - 52
outside
53 - 61
TMhelix
62 - 84
inside
85 - 104
TMhelix
105 - 127
outside
128 - 139
TMhelix
140 - 162
inside
163 - 227
Population Genetic Test Statistics
Pi
132.254533
Theta
150.856395
Tajima's D
-0.413757
CLR
0.099585
CSRT
0.267486625668717
Interpretation
Uncertain
