Pre Gene Modal
BGIBMGA007247
Annotation
PREDICTED:_nuclear_distribution_protein_nudE-like_1_isoform_X1_[Bombyx_mori]
Full name
Nuclear distribution protein nudE-like 1-B
+ More
Nuclear distribution protein nudE homolog 1
Nuclear distribution protein nudE homolog 1
Location in the cell
Nuclear Reliability : 3.657
Sequence
CDS
ATGCAATCTCCGAAACAAGCAGATTGTGACACAGTGGAGTACTGGAAAAATCTAGCAGAATGCTATGAACAGAAAGTGAACGACATTCAGCAAGAATTAGATGAATACACAGAAAATTCGGCTCAGTTGGAAAAGGAATTGGAAGCATCATTAGGGCAAGTTGAAAAGCAGAACAGAGATCTGGAACACCAAAATCAACGCCTTAAAAATGAAATTGAAATGTTAAAAAACAAGCTGGAAAGAAGTCAATATGAGACTAATGCTTTAGAAAATGAACTACAAGCTTTAAAGATAGATAAGGACAAGCAGGCAGCATATGTGAGGGAATTGGAACAAAAGAACGATGATCTGGAAAGAGGTCAAAGAGTCATATCAGAATCCGTATCGTGTATAGAAACAATGTTAAATGAAGCATATGAGAGGAATGCAGTTCTTGAAAATGAAGTTGATGAAATAGAAAATCTTAGGATAAAATTGCAGAGAGCAAATGATGAAGCTAGAGATCTGAAACAAGAATTAAAAGTTGTTGAAAGAATTCCGTCAAGTAAGAAAGAGGAAGATGAAAATGAAACAACATTAACGAACGGACATCTTTCAAATTCAACTCGGAATCAAGTTGAGATTGAGACACAAACATCCATTATGTCACCACAAAAACGGGAAATAAATGGTAACGCAATGACACCATCGGCCAGAGTTTCAGCAATAAATTTAGTTGGTGATCTCCTTAGGAAGGTTGGGGCTTTGGAATCGAAATTAGCATCGTGTCGTGGTACAGTTAGACCTAAAGAGCATTCACCAAACACGACCTCGGAAGTCAATAAGGACTACAGATGTGTACTCAAGAATTAA
Protein
MQSPKQADCDTVEYWKNLAECYEQKVNDIQQELDEYTENSAQLEKELEASLGQVEKQNRDLEHQNQRLKNEIEMLKNKLERSQYETNALENELQALKIDKDKQAAYVRELEQKNDDLERGQRVISESVSCIETMLNEAYERNAVLENEVDEIENLRIKLQRANDEARDLKQELKVVERIPSSKKEEDENETTLTNGHLSNSTRNQVEIETQTSIMSPQKREINGNAMTPSARVSAINLVGDLLRKVGALESKLASCRGTVRPKEHSPNTTSEVNKDYRCVLKN
Summary
Description
Required for organization of the cellular microtubule array and microtubule anchoring at the centrosome. Positively regulates the activity of the minus-end directed microtubule motor protein dynein. May enhance dynein-mediated microtubule sliding by targeting dynein to the microtubule plus end (By similarity).
Required for centrosome duplication and formation and function of the mitotic spindle. Essential for the development of the cerebral cortex. May regulate the production of neurons by controlling the orientation of the mitotic spindle during division of cortical neuronal progenitors of the proliferative ventricular zone of the brain. Orientation of the division plane perpendicular to the layers of the cortex gives rise to two proliferative neuronal progenitors whereas parallel orientation of the division plane yields one proliferative neuronal progenitor and a post-mitotic neuron. A premature shift towards a neuronal fate within the progenitor population may result in an overall reduction in the final number of neurons and an increase in the number of neurons in the deeper layers of the cortex.
Required for centrosome duplication and formation and function of the mitotic spindle. Essential for the development of the cerebral cortex. May regulate the production of neurons by controlling the orientation of the mitotic spindle during division of cortical neuronal progenitors of the proliferative ventricular zone of the brain. Orientation of the division plane perpendicular to the layers of the cortex gives rise to two proliferative neuronal progenitors whereas parallel orientation of the division plane yields one proliferative neuronal progenitor and a post-mitotic neuron. A premature shift towards a neuronal fate within the progenitor population may result in an overall reduction in the final number of neurons and an increase in the number of neurons in the deeper layers of the cortex.
Subunit
Self-associates. Interacts with CNTRL, LIS1, dynein, SLMAP and TCP1 (By similarity). Interacts with CENPF, dynactin, tubulin gamma, PAFAH1B1, PCM1 and PCNT. Interacts with ZNF365.
Similarity
Belongs to the nudE family.
Keywords
Coiled coil
Complete proteome
Cytoplasm
Cytoskeleton
Microtubule
Phosphoprotein
Reference proteome
Transport
Alternative splicing
Cell cycle
Cell division
Centromere
Chromosome
Developmental protein
Differentiation
Kinetochore
Lipoprotein
Lissencephaly
Mitosis
Neurogenesis
Palmitate
Feature
chain Nuclear distribution protein nudE-like 1-B
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JCK2
A0A3S2P3A2
A0A2A4JU80
A0A194QCV7
A0A194QX21
A0A212F6H6
+ More
A0A2D0Q421 K9III5 A0A3B1K1X6 A0A3Q0RSN3 A0A3B4EG99 A0A3Q3BNM0 A0A3P9D3G5 A0A3P8QYJ8 A0A3B4FPM0 A0A3Q1JNV9 A0A3Q7SEE4 A0A2K6SFX6 A7SPT5 G5AK09 F6X6L7 A0A3Q2HSQ5 L5M6F0 A0A2K6SFU9 A0A2K6SFU7 A0A287BHJ3 S7PS52 A0A3Q7MU00 F6WM97 A0A091QFY1 I3L8G0 U6CS63 A0A3Q7P6W8 A0A287AZB1 A0A2U3XB89 A0A2Y9HX88 G3TE08 A0A3Q2KLE7 F7CL14 A0A2K5PNP9 A0A340Y3L2 A0A341BAY5 A0A2Y9LER2 A0A2U4ATB0 A0A2Y9LLZ9 A0A341BBB3 A0A2Y9LRY1 A0A2Y9KFB8 A0A341BEG3 A0A2Y9LH55 A0A3Q7Y9M5 A0A2U4AT96 H0WN99 A0A341BCW5 A0A2Y9LH50 A0A2Y9LEH7 A0A3Q7WT80 A0A060W9B3 A0A2Y9LEQ7 M3YS77 G1SD04 L9KXL8 A0A2Y9KSQ5 A0A2K5PNL9 A0A093SVZ7 A0A093NBL7 A0A2K5PNL5 B2GRS6 Q803Q2 A0A384BVE7 A0A3Q7WVA5 A0A2K6DJ70 A0A2K5KMY0 A0A1U7SL92 A0A2J8IPJ6 A0A093ICX5 A0A2K6JR18 A0A2K6PFW5 F7AU88 A0A2K5KN04 A0A093C364 A0A2K5KMX3 A0A096NLV7 G3W7T6 A0A250YFD2 A0A2K6DJ87 G7Q0J8 A0A3Q2DNZ5 A0A2K6JR41 A0A2K6PFW8 A0A3Q0EC48 A0A091NI17 Q9NXR1-1 A0A384CJX1 U6CUR5 A0A3Q7SWD1 A0A2U3VTA3 A0A2Y9HF52 A0A3Q7NFV8
A0A2D0Q421 K9III5 A0A3B1K1X6 A0A3Q0RSN3 A0A3B4EG99 A0A3Q3BNM0 A0A3P9D3G5 A0A3P8QYJ8 A0A3B4FPM0 A0A3Q1JNV9 A0A3Q7SEE4 A0A2K6SFX6 A7SPT5 G5AK09 F6X6L7 A0A3Q2HSQ5 L5M6F0 A0A2K6SFU9 A0A2K6SFU7 A0A287BHJ3 S7PS52 A0A3Q7MU00 F6WM97 A0A091QFY1 I3L8G0 U6CS63 A0A3Q7P6W8 A0A287AZB1 A0A2U3XB89 A0A2Y9HX88 G3TE08 A0A3Q2KLE7 F7CL14 A0A2K5PNP9 A0A340Y3L2 A0A341BAY5 A0A2Y9LER2 A0A2U4ATB0 A0A2Y9LLZ9 A0A341BBB3 A0A2Y9LRY1 A0A2Y9KFB8 A0A341BEG3 A0A2Y9LH55 A0A3Q7Y9M5 A0A2U4AT96 H0WN99 A0A341BCW5 A0A2Y9LH50 A0A2Y9LEH7 A0A3Q7WT80 A0A060W9B3 A0A2Y9LEQ7 M3YS77 G1SD04 L9KXL8 A0A2Y9KSQ5 A0A2K5PNL9 A0A093SVZ7 A0A093NBL7 A0A2K5PNL5 B2GRS6 Q803Q2 A0A384BVE7 A0A3Q7WVA5 A0A2K6DJ70 A0A2K5KMY0 A0A1U7SL92 A0A2J8IPJ6 A0A093ICX5 A0A2K6JR18 A0A2K6PFW5 F7AU88 A0A2K5KN04 A0A093C364 A0A2K5KMX3 A0A096NLV7 G3W7T6 A0A250YFD2 A0A2K6DJ87 G7Q0J8 A0A3Q2DNZ5 A0A2K6JR41 A0A2K6PFW8 A0A3Q0EC48 A0A091NI17 Q9NXR1-1 A0A384CJX1 U6CUR5 A0A3Q7SWD1 A0A2U3VTA3 A0A2Y9HF52 A0A3Q7NFV8
Pubmed
19121390
26354079
22118469
25329095
25186727
17615350
+ More
21993625 16341006 19892987 17495919 24755649 21993624 23385571 25362486 21709235 28087693 22002653 14702039 15616553 15489334 12556484 16291865 16682949 17600710 18220336 18669648 19927128 20068231 21529752 21529751 22526350 23186163 24275569 24813606
21993625 16341006 19892987 17495919 24755649 21993624 23385571 25362486 21709235 28087693 22002653 14702039 15616553 15489334 12556484 16291865 16682949 17600710 18220336 18669648 19927128 20068231 21529752 21529751 22526350 23186163 24275569 24813606
EMBL
BABH01039082
RSAL01000036
RVE51262.1
NWSH01000569
PCG75587.1
KQ459232
+ More
KPJ02815.1 KQ461155 KPJ08121.1 AGBW02010016 OWR49333.1 GABZ01006835 JAA46690.1 DS469737 EDO34270.1 JH165553 GEBF01003213 EHA97369.1 JAO00420.1 AAEX03004498 KB104461 ELK33288.1 AEMK02000014 KE163235 EPQ11317.1 KK698649 KFQ25925.1 HAAF01001333 CCP73159.1 AAQR03077633 AAQR03077634 AAQR03077635 FR904390 CDQ61814.1 AEYP01027378 AEYP01027379 AEYP01027380 AEYP01027381 AAGW02057154 AAGW02057155 AAGW02057156 KB320604 ELW67680.1 KL672660 KFW86684.1 KL224628 KFW62343.1 BC165239 AAI65239.1 BC044386 NBAG03000658 PNI12438.1 KK591609 KFW00530.1 KL232548 KFV06697.1 AHZZ02018409 AEFK01165078 AEFK01165079 AEFK01165080 AEFK01165081 AEFK01165082 AEFK01165083 AEFK01165084 AEFK01165085 AEFK01165086 AEFK01165087 GFFW01002441 JAV42347.1 CM001295 EHH60186.1 KL383607 KFP89111.1 AK000108 AC026401 AF001548 BC001421 BC033900 HAAF01002390 CCP74216.1
KPJ02815.1 KQ461155 KPJ08121.1 AGBW02010016 OWR49333.1 GABZ01006835 JAA46690.1 DS469737 EDO34270.1 JH165553 GEBF01003213 EHA97369.1 JAO00420.1 AAEX03004498 KB104461 ELK33288.1 AEMK02000014 KE163235 EPQ11317.1 KK698649 KFQ25925.1 HAAF01001333 CCP73159.1 AAQR03077633 AAQR03077634 AAQR03077635 FR904390 CDQ61814.1 AEYP01027378 AEYP01027379 AEYP01027380 AEYP01027381 AAGW02057154 AAGW02057155 AAGW02057156 KB320604 ELW67680.1 KL672660 KFW86684.1 KL224628 KFW62343.1 BC165239 AAI65239.1 BC044386 NBAG03000658 PNI12438.1 KK591609 KFW00530.1 KL232548 KFV06697.1 AHZZ02018409 AEFK01165078 AEFK01165079 AEFK01165080 AEFK01165081 AEFK01165082 AEFK01165083 AEFK01165084 AEFK01165085 AEFK01165086 AEFK01165087 GFFW01002441 JAV42347.1 CM001295 EHH60186.1 KL383607 KFP89111.1 AK000108 AC026401 AF001548 BC001421 BC033900 HAAF01002390 CCP74216.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000221080 UP000018467 UP000261340 UP000261440 UP000264840 UP000265160 UP000265100 UP000261460 UP000265040 UP000286640 UP000233220 UP000001593 UP000006813 UP000002254 UP000002281 UP000008227 UP000286641 UP000245341 UP000248481 UP000007646 UP000002280 UP000233040 UP000265300 UP000252040 UP000248483 UP000245320 UP000248482 UP000286642 UP000005225 UP000193380 UP000000715 UP000001811 UP000011518 UP000053258 UP000054081 UP000000437 UP000261680 UP000233120 UP000233060 UP000189705 UP000233180 UP000233200 UP000008225 UP000028761 UP000007648 UP000009130 UP000265020 UP000189704 UP000005640 UP000291021 UP000245340
UP000221080 UP000018467 UP000261340 UP000261440 UP000264840 UP000265160 UP000265100 UP000261460 UP000265040 UP000286640 UP000233220 UP000001593 UP000006813 UP000002254 UP000002281 UP000008227 UP000286641 UP000245341 UP000248481 UP000007646 UP000002280 UP000233040 UP000265300 UP000252040 UP000248483 UP000245320 UP000248482 UP000286642 UP000005225 UP000193380 UP000000715 UP000001811 UP000011518 UP000053258 UP000054081 UP000000437 UP000261680 UP000233120 UP000233060 UP000189705 UP000233180 UP000233200 UP000008225 UP000028761 UP000007648 UP000009130 UP000265020 UP000189704 UP000005640 UP000291021 UP000245340
Pfam
PF04880 NUDE_C
ProteinModelPortal
H9JCK2
A0A3S2P3A2
A0A2A4JU80
A0A194QCV7
A0A194QX21
A0A212F6H6
+ More
A0A2D0Q421 K9III5 A0A3B1K1X6 A0A3Q0RSN3 A0A3B4EG99 A0A3Q3BNM0 A0A3P9D3G5 A0A3P8QYJ8 A0A3B4FPM0 A0A3Q1JNV9 A0A3Q7SEE4 A0A2K6SFX6 A7SPT5 G5AK09 F6X6L7 A0A3Q2HSQ5 L5M6F0 A0A2K6SFU9 A0A2K6SFU7 A0A287BHJ3 S7PS52 A0A3Q7MU00 F6WM97 A0A091QFY1 I3L8G0 U6CS63 A0A3Q7P6W8 A0A287AZB1 A0A2U3XB89 A0A2Y9HX88 G3TE08 A0A3Q2KLE7 F7CL14 A0A2K5PNP9 A0A340Y3L2 A0A341BAY5 A0A2Y9LER2 A0A2U4ATB0 A0A2Y9LLZ9 A0A341BBB3 A0A2Y9LRY1 A0A2Y9KFB8 A0A341BEG3 A0A2Y9LH55 A0A3Q7Y9M5 A0A2U4AT96 H0WN99 A0A341BCW5 A0A2Y9LH50 A0A2Y9LEH7 A0A3Q7WT80 A0A060W9B3 A0A2Y9LEQ7 M3YS77 G1SD04 L9KXL8 A0A2Y9KSQ5 A0A2K5PNL9 A0A093SVZ7 A0A093NBL7 A0A2K5PNL5 B2GRS6 Q803Q2 A0A384BVE7 A0A3Q7WVA5 A0A2K6DJ70 A0A2K5KMY0 A0A1U7SL92 A0A2J8IPJ6 A0A093ICX5 A0A2K6JR18 A0A2K6PFW5 F7AU88 A0A2K5KN04 A0A093C364 A0A2K5KMX3 A0A096NLV7 G3W7T6 A0A250YFD2 A0A2K6DJ87 G7Q0J8 A0A3Q2DNZ5 A0A2K6JR41 A0A2K6PFW8 A0A3Q0EC48 A0A091NI17 Q9NXR1-1 A0A384CJX1 U6CUR5 A0A3Q7SWD1 A0A2U3VTA3 A0A2Y9HF52 A0A3Q7NFV8
A0A2D0Q421 K9III5 A0A3B1K1X6 A0A3Q0RSN3 A0A3B4EG99 A0A3Q3BNM0 A0A3P9D3G5 A0A3P8QYJ8 A0A3B4FPM0 A0A3Q1JNV9 A0A3Q7SEE4 A0A2K6SFX6 A7SPT5 G5AK09 F6X6L7 A0A3Q2HSQ5 L5M6F0 A0A2K6SFU9 A0A2K6SFU7 A0A287BHJ3 S7PS52 A0A3Q7MU00 F6WM97 A0A091QFY1 I3L8G0 U6CS63 A0A3Q7P6W8 A0A287AZB1 A0A2U3XB89 A0A2Y9HX88 G3TE08 A0A3Q2KLE7 F7CL14 A0A2K5PNP9 A0A340Y3L2 A0A341BAY5 A0A2Y9LER2 A0A2U4ATB0 A0A2Y9LLZ9 A0A341BBB3 A0A2Y9LRY1 A0A2Y9KFB8 A0A341BEG3 A0A2Y9LH55 A0A3Q7Y9M5 A0A2U4AT96 H0WN99 A0A341BCW5 A0A2Y9LH50 A0A2Y9LEH7 A0A3Q7WT80 A0A060W9B3 A0A2Y9LEQ7 M3YS77 G1SD04 L9KXL8 A0A2Y9KSQ5 A0A2K5PNL9 A0A093SVZ7 A0A093NBL7 A0A2K5PNL5 B2GRS6 Q803Q2 A0A384BVE7 A0A3Q7WVA5 A0A2K6DJ70 A0A2K5KMY0 A0A1U7SL92 A0A2J8IPJ6 A0A093ICX5 A0A2K6JR18 A0A2K6PFW5 F7AU88 A0A2K5KN04 A0A093C364 A0A2K5KMX3 A0A096NLV7 G3W7T6 A0A250YFD2 A0A2K6DJ87 G7Q0J8 A0A3Q2DNZ5 A0A2K6JR41 A0A2K6PFW8 A0A3Q0EC48 A0A091NI17 Q9NXR1-1 A0A384CJX1 U6CUR5 A0A3Q7SWD1 A0A2U3VTA3 A0A2Y9HF52 A0A3Q7NFV8
PDB
2V71
E-value=2.28806e-22,
Score=259
Ontologies
GO
GO:2000574
GO:0007399
GO:0060052
GO:0007100
GO:0007020
GO:0045202
GO:0042802
GO:0051298
GO:0008017
GO:0000776
GO:0007405
GO:0047496
GO:0000132
GO:0031616
GO:0001764
GO:0051303
GO:0021987
GO:0051642
GO:0005813
GO:0016477
GO:0005871
GO:0007059
GO:0032418
GO:0090630
GO:0021955
GO:1990138
GO:1900029
GO:0043203
GO:0008090
GO:0021799
GO:0031252
GO:0043014
GO:0048487
GO:0060053
GO:0001833
GO:0010975
GO:0008021
GO:0033157
GO:0008286
GO:1904115
GO:0005874
GO:0005819
GO:0005737
GO:0005875
GO:0010389
GO:0000777
GO:0000086
GO:0005829
GO:0032154
GO:0097711
GO:0019904
GO:0016020
GO:0051301
PANTHER
Topology
Subcellular location
Cytoplasm
Localizes to the interphase centrosome and the mitotic spindle. With evidence from 1 publications.
Cytoskeleton Localizes to the interphase centrosome and the mitotic spindle. With evidence from 1 publications.
Microtubule organizing center Localizes to the interphase centrosome and the mitotic spindle. With evidence from 1 publications.
Centrosome Localizes to the interphase centrosome and the mitotic spindle. With evidence from 1 publications.
Spindle Localizes to the interphase centrosome and the mitotic spindle. With evidence from 1 publications.
Chromosome
Centromere
Kinetochore
Cleavage furrow
Cytoskeleton Localizes to the interphase centrosome and the mitotic spindle. With evidence from 1 publications.
Microtubule organizing center Localizes to the interphase centrosome and the mitotic spindle. With evidence from 1 publications.
Centrosome Localizes to the interphase centrosome and the mitotic spindle. With evidence from 1 publications.
Spindle Localizes to the interphase centrosome and the mitotic spindle. With evidence from 1 publications.
Chromosome
Centromere
Kinetochore
Cleavage furrow
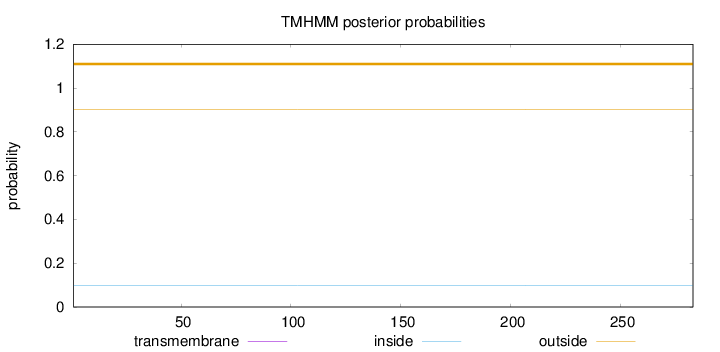
Length:
283
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00039
Exp number, first 60 AAs:
0
Total prob of N-in:
0.09708
outside
1 - 283
Population Genetic Test Statistics
Pi
212.731969
Theta
179.927267
Tajima's D
0.463238
CLR
0
CSRT
0.502124893755312
Interpretation
Uncertain
