Gene
KWMTBOMO10342
Pre Gene Modal
BGIBMGA007228
Annotation
potassium_coupled_amino_acid_transporter_[Manduca_sexta]
Full name
Transporter
+ More
Sodium-dependent nutrient amino acid transporter 1
Sodium-dependent nutrient amino acid transporter 1
Location in the cell
PlasmaMembrane Reliability : 4.949
Sequence
CDS
ATGAGTCAAACACATGTGAACAATGGGTTCGACATGTCCTCGGAGAATCCTCCGACGGACAGGGACCCGTCATTGAATCGTCCAGGATCACCCAAGCCGAAGCGATCCACAAAACCTGATCTACCTCCTCCGGCACTGAAGGTTCCAGTCAATGAAAAGAAGGCTTTCGAAGATATAGAAGGTGCTCCTGAACGTATGATGTGGTCTAATCAAATTGAATTCCTCATGTCGTGCATCGCTACATCAGTGGGCTTAGGCAACGTATGGCGGTTTCCATTCGTTGCATATCAAAATGGCGGCGGGGCCTTCCTTATACCCTATATCATAGTGTTGTTTTTAGTCGGCAAACCCATGTACTACCTCGAATGTGTTTTGGGACAGTTTAGTTCGAGAAATTCTGTCAAAGTTTGGTCTTTATCACCAGCAATGAAAGGAACCGGCTATGCTCAAGCTTTGGGTTGCGGATACATCCTATCTTACTACGTAGTAATTGTTGGTCTATGCCTGTATTATTTGTCAATGAGTTTCCAGTCTACACTGCCATGGGCTGTTTGTGAACCTGGTTGGGAAAACTGCGTGCCTTCGGATCCTAACAGTGCCCCAGTTGGAACCGTAGTCAACGGCACCAGTAGCGCGGAGCTTTACTTCTTGAGAACAGTTCTTCAGCAAAGCGATGGTATCGAAGGCGGACTCGGCGCTCCTATATGGTACCTGACCTTATGTTTGCTCGCAACGTGGGTGATTATTTTTGTCATCGTCGCACAAGGCGTGAAGAGTTCTGGCAAGGCTGCATACTTCCTAGCTTTGTTCCCCTATGTCGTTATGATCATATTGTTCATAACGACTGTAATTTTACCCGGAGCTGGTACGGGAATCCTCTTCTTCATAACGCCTCAATGGAGTAAACTATTAGAGTTAGACGTATGGTATGCTGCAGTAACGCAAGTATTCTTCTCGTTAACAGTTTGCACTGGAGCTATCATCATGTTCTCATCCTATAATGGTTTCAAACAAAATGTCTACAGAGATGCAATGATTGTTACAACTTTGGACACCCTCACAAGTTTGCTATCCGGCTTTACTATTTTCGGTATCCTCGGTAACTTGGCTAATGAACTAAATTCTCGAGTAGAAGATGTGGTCGGCTCTGGAGGAACCGGACTAGCTTTCATCTCGTACCCTGACGCCATTGCAAAAACGTTCATGCCACAGTTGTTCTCAGTTCTGTTCTTCCTGATGATGGCTGTGCTGGGTGTGGGTTCATCTGTGGCTCTTTTGTCGACCATCAACACTGTAGTGATGGACGCTTTCCCCAACGTTCCCACAGTGTTCATGTCGGCTCTCTGCTGTACCGTTGGATTTTTAGTGGGACTTGTTTACGTCACACCTGGTGGACAATATATATTGGAGCTTGTCGATTACTACGGTGGAACGTTTTTAGTATTATTCTGCGCCATTGCTGAAATTATTGGTGTATTCTGGATATACGGCTTAGAGAATCTGTGCCTCGACATCGAATACATGTTGGATATAAAGACGTCTCCTTACTGGCGCATGTGTTGGGGTCTGATCACGCCAGCCATGATGATTATTGTATTCCTGTACGCACTGATTTCATACGAAGCATTGCTCTTCGGAGGAATGGCCGGATATGTGGCTGGTTACCTGTTGTTGATCGTGGGTATTTTGTTGGTACCGATTTCCATCTGCCTTACATTGTACAAGTACAGGACTGGAAACTTCATTGAGACTCTAAAGCTTTCTTTCCGCTCCAAGGAATCTTGGGGTCCAAGATCAGCTGGCGAACGCAGAACCTGGCTTTCATTTAAAGAAGACGCGAAAGTATTACGAGAAAAACTTAATCCCTCACGAATCACGCATGTCTGGTATAGTTTAATAGGAAGCTACAGACGACGAATCTAA
Protein
MSQTHVNNGFDMSSENPPTDRDPSLNRPGSPKPKRSTKPDLPPPALKVPVNEKKAFEDIEGAPERMMWSNQIEFLMSCIATSVGLGNVWRFPFVAYQNGGGAFLIPYIIVLFLVGKPMYYLECVLGQFSSRNSVKVWSLSPAMKGTGYAQALGCGYILSYYVVIVGLCLYYLSMSFQSTLPWAVCEPGWENCVPSDPNSAPVGTVVNGTSSAELYFLRTVLQQSDGIEGGLGAPIWYLTLCLLATWVIIFVIVAQGVKSSGKAAYFLALFPYVVMIILFITTVILPGAGTGILFFITPQWSKLLELDVWYAAVTQVFFSLTVCTGAIIMFSSYNGFKQNVYRDAMIVTTLDTLTSLLSGFTIFGILGNLANELNSRVEDVVGSGGTGLAFISYPDAIAKTFMPQLFSVLFFLMMAVLGVGSSVALLSTINTVVMDAFPNVPTVFMSALCCTVGFLVGLVYVTPGGQYILELVDYYGGTFLVLFCAIAEIIGVFWIYGLENLCLDIEYMLDIKTSPYWRMCWGLITPAMMIIVFLYALISYEALLFGGMAGYVAGYLLLIVGILLVPISICLTLYKYRTGNFIETLKLSFRSKESWGPRSAGERRTWLSFKEDAKVLREKLNPSRITHVWYSLIGSYRRRI
Summary
Description
Unusual broad substrate spectrum amino acid:sodium cotransporter that promotes absorption of the D isomers of essential amino acids. Neutral amino acids are the preferred substrates, especially methionine and phenylalanine (By similarity).
Unusual broad substrate spectrum amino acid:sodium cotransporter that promotes absorption of the D isomers of essential amino acids. Neutral amino acids are the preferred substrates, especially methionine and phenylalanine.
Unusual broad substrate spectrum amino acid:sodium cotransporter that promotes absorption of the D isomers of essential amino acids. Neutral amino acids are the preferred substrates, especially methionine and phenylalanine.
Similarity
Belongs to the sodium:neurotransmitter symporter (SNF) (TC 2.A.22) family.
Keywords
Amino-acid transport
Complete proteome
Glycoprotein
Ion transport
Membrane
Sodium
Sodium transport
Symport
Transmembrane
Transmembrane helix
Transport
Reference proteome
Feature
chain Sodium-dependent nutrient amino acid transporter 1
Uniprot
H9JCI3
O76188
Q9U5A9
A0A194QSZ7
A0A194QH91
A0A2H1VRW7
+ More
A0A2A4JUB4 A0A2A4JNI2 A0A1E1VZ02 B2ZXL8 A0A3S2M4K9 A0A2A4JN91 A0A2A4JPM7 A0A2A4JQ43 A0A2A4JQ17 A0A2H1VSW1 A0A212EKM3 A0A1E1WV88 A0A1E1W207 A0A212EKP1 A0A2J7PKX9 A0A1I8Q6E3 A0A2J7PKT0 A0A088AJX5 A0A0L7RF06 A0A310SK53 A0A1Q3G5A3 A0A232F021 Q6VS78 E0VLW4 B3NV41 Q16YU3 A0A1B0B655 J9HFX8 Q8IST0 A0A1A9XLV3 A0A1A9WAV7 A0A067RFW9 A0A0K8TP80 U5EJQ1 A0A195DL34 A0A1I8MYH7 A0A1J1HQJ1 B4NDL8 B4GVM9 B4L7U0 D6WWF3 A0A310S8Y0 A0A3L8DNA2 B4PZQ4 B4R4T6 A0A088AJW4 A0A0L0BU43 B4MEG2 A0A3B0JYF6 A0A2A3ECG5 A0A088AJX6 Q29GB8 C7LA87 A0A2A3EB77 A0A0A1XSM6 Q9W4C5 A0A1W4V6M6 A0A336LX44 A0A1B0AE48 A0A3L8DT63 A0A1A9UT47 E2AWU5 E2BSN7 A0A158NLY9 A0A1B0B654 A0A0K8V9C6 A0A336KAS2 A0A0P6GKY2 A0A1L8DK96 A0A1L8DJX3 A0A182GFI3 A0A0P5G743 A0A336KF85 A0A1B0DCW8 A0A151XF32 A0A0P6HGT0 A0A1L8DJX4 A0A0N8A3A2 B5DZG7 A0A0M9ABU2 A0A0R3NLF5 A0A0M4E615 A0A3B0J223 A0A0P5RG87 E9G979 A0A034WJ71 A0A182GIY5 A0A0A1XHG2 A0A3B0JVN4 B4GDP4 A0A0M4EX99 A0A158NLY7
A0A2A4JUB4 A0A2A4JNI2 A0A1E1VZ02 B2ZXL8 A0A3S2M4K9 A0A2A4JN91 A0A2A4JPM7 A0A2A4JQ43 A0A2A4JQ17 A0A2H1VSW1 A0A212EKM3 A0A1E1WV88 A0A1E1W207 A0A212EKP1 A0A2J7PKX9 A0A1I8Q6E3 A0A2J7PKT0 A0A088AJX5 A0A0L7RF06 A0A310SK53 A0A1Q3G5A3 A0A232F021 Q6VS78 E0VLW4 B3NV41 Q16YU3 A0A1B0B655 J9HFX8 Q8IST0 A0A1A9XLV3 A0A1A9WAV7 A0A067RFW9 A0A0K8TP80 U5EJQ1 A0A195DL34 A0A1I8MYH7 A0A1J1HQJ1 B4NDL8 B4GVM9 B4L7U0 D6WWF3 A0A310S8Y0 A0A3L8DNA2 B4PZQ4 B4R4T6 A0A088AJW4 A0A0L0BU43 B4MEG2 A0A3B0JYF6 A0A2A3ECG5 A0A088AJX6 Q29GB8 C7LA87 A0A2A3EB77 A0A0A1XSM6 Q9W4C5 A0A1W4V6M6 A0A336LX44 A0A1B0AE48 A0A3L8DT63 A0A1A9UT47 E2AWU5 E2BSN7 A0A158NLY9 A0A1B0B654 A0A0K8V9C6 A0A336KAS2 A0A0P6GKY2 A0A1L8DK96 A0A1L8DJX3 A0A182GFI3 A0A0P5G743 A0A336KF85 A0A1B0DCW8 A0A151XF32 A0A0P6HGT0 A0A1L8DJX4 A0A0N8A3A2 B5DZG7 A0A0M9ABU2 A0A0R3NLF5 A0A0M4E615 A0A3B0J223 A0A0P5RG87 E9G979 A0A034WJ71 A0A182GIY5 A0A0A1XHG2 A0A3B0JVN4 B4GDP4 A0A0M4EX99 A0A158NLY7
Pubmed
EMBL
BABH01039076
BABH01039077
BABH01039078
BABH01039079
AF006063
AAC24190.1
+ More
AF013963 AAF18560.1 KQ461155 KPJ08125.1 KQ459232 KPJ02811.1 ODYU01004070 SOQ43560.1 NWSH01000569 PCG75591.1 NWSH01000921 PCG73531.1 GDQN01011140 JAT79914.1 AB365597 BAG39453.1 RSAL01000036 RVE51256.1 PCG73535.1 PCG73534.1 PCG73532.1 PCG73533.1 SOQ43562.1 AGBW02014242 OWR42034.1 GDQN01004191 GDQN01000328 JAT86863.1 JAT90726.1 GDQN01010095 JAT80959.1 OWR42035.1 NEVH01024531 PNF16994.1 PNF16938.1 KQ414608 KOC69394.1 KQ763581 OAD54951.1 GFDL01000068 JAV34977.1 NNAY01001443 OXU23962.1 AY330296 AAR08269.1 DS235282 EEB14370.1 CH954180 EDV45889.1 CH477509 EAT39788.1 JXJN01008983 EJY57747.1 AF543193 AAN40410.1 KK852489 KDR22766.1 GDAI01001449 JAI16154.1 GANO01002161 JAB57710.1 KQ980762 KYN13552.1 CVRI01000017 CRK90243.1 CH964239 EDW81840.1 CH479193 EDW26724.1 CH933814 EDW05515.1 KQ971361 EFA08708.1 OAD54950.1 QOIP01000006 RLU21298.1 CM000162 EDX01121.1 CM000366 EDX17133.1 JRES01001335 KNC23567.1 CH940664 EDW62937.1 OUUW01000003 SPP78406.1 KZ288308 PBC28721.1 CH379064 EAL32192.1 BT099637 ACU68946.1 PBC28724.1 GBXI01000417 JAD13875.1 DQ073009 AE014298 AY121662 AAF46031.2 AAM51989.1 UFQS01000248 UFQT01000248 SSX01993.1 SSX22370.1 QOIP01000004 RLU23635.1 GL443425 EFN62065.1 GL450241 EFN81283.1 ADTU01020082 ADTU01020083 ADTU01020084 ADTU01020085 ADTU01020086 ADTU01020087 ADTU01020088 ADTU01020089 ADTU01020090 ADTU01020091 JXJN01008984 GDHF01016852 JAI35462.1 SSX01994.1 SSX22371.1 GDIQ01033961 JAN60776.1 GFDF01007320 JAV06764.1 GFDF01007322 JAV06762.1 JXUM01010513 KQ560310 KXJ83117.1 GDIQ01245340 JAK06385.1 SSX01995.1 SSX22372.1 AJVK01014254 KQ982205 KYQ59002.1 GDIQ01028602 JAN66135.1 GFDF01007321 JAV06763.1 GDIP01186721 LRGB01001348 JAJ36681.1 KZS12914.1 CM000071 EDY68808.1 KQ435689 KOX81270.1 KRT01700.1 CP012524 ALC41841.1 OUUW01000001 SPP75127.1 GDIQ01101096 JAL50630.1 GL732535 EFX84094.1 GAKP01004550 JAC54402.1 JXUM01067031 JXUM01067032 JXUM01067033 JXUM01067034 JXUM01067035 JXUM01067036 KQ562429 KXJ75930.1 GBXI01003895 JAD10397.1 SPP75128.1 CH479181 EDW31701.1 CP012528 ALC48305.1 ADTU01019952 ADTU01019953 ADTU01019954 ADTU01019955 ADTU01019956 ADTU01019957 ADTU01019958 ADTU01019959 ADTU01019960 ADTU01019961
AF013963 AAF18560.1 KQ461155 KPJ08125.1 KQ459232 KPJ02811.1 ODYU01004070 SOQ43560.1 NWSH01000569 PCG75591.1 NWSH01000921 PCG73531.1 GDQN01011140 JAT79914.1 AB365597 BAG39453.1 RSAL01000036 RVE51256.1 PCG73535.1 PCG73534.1 PCG73532.1 PCG73533.1 SOQ43562.1 AGBW02014242 OWR42034.1 GDQN01004191 GDQN01000328 JAT86863.1 JAT90726.1 GDQN01010095 JAT80959.1 OWR42035.1 NEVH01024531 PNF16994.1 PNF16938.1 KQ414608 KOC69394.1 KQ763581 OAD54951.1 GFDL01000068 JAV34977.1 NNAY01001443 OXU23962.1 AY330296 AAR08269.1 DS235282 EEB14370.1 CH954180 EDV45889.1 CH477509 EAT39788.1 JXJN01008983 EJY57747.1 AF543193 AAN40410.1 KK852489 KDR22766.1 GDAI01001449 JAI16154.1 GANO01002161 JAB57710.1 KQ980762 KYN13552.1 CVRI01000017 CRK90243.1 CH964239 EDW81840.1 CH479193 EDW26724.1 CH933814 EDW05515.1 KQ971361 EFA08708.1 OAD54950.1 QOIP01000006 RLU21298.1 CM000162 EDX01121.1 CM000366 EDX17133.1 JRES01001335 KNC23567.1 CH940664 EDW62937.1 OUUW01000003 SPP78406.1 KZ288308 PBC28721.1 CH379064 EAL32192.1 BT099637 ACU68946.1 PBC28724.1 GBXI01000417 JAD13875.1 DQ073009 AE014298 AY121662 AAF46031.2 AAM51989.1 UFQS01000248 UFQT01000248 SSX01993.1 SSX22370.1 QOIP01000004 RLU23635.1 GL443425 EFN62065.1 GL450241 EFN81283.1 ADTU01020082 ADTU01020083 ADTU01020084 ADTU01020085 ADTU01020086 ADTU01020087 ADTU01020088 ADTU01020089 ADTU01020090 ADTU01020091 JXJN01008984 GDHF01016852 JAI35462.1 SSX01994.1 SSX22371.1 GDIQ01033961 JAN60776.1 GFDF01007320 JAV06764.1 GFDF01007322 JAV06762.1 JXUM01010513 KQ560310 KXJ83117.1 GDIQ01245340 JAK06385.1 SSX01995.1 SSX22372.1 AJVK01014254 KQ982205 KYQ59002.1 GDIQ01028602 JAN66135.1 GFDF01007321 JAV06763.1 GDIP01186721 LRGB01001348 JAJ36681.1 KZS12914.1 CM000071 EDY68808.1 KQ435689 KOX81270.1 KRT01700.1 CP012524 ALC41841.1 OUUW01000001 SPP75127.1 GDIQ01101096 JAL50630.1 GL732535 EFX84094.1 GAKP01004550 JAC54402.1 JXUM01067031 JXUM01067032 JXUM01067033 JXUM01067034 JXUM01067035 JXUM01067036 KQ562429 KXJ75930.1 GBXI01003895 JAD10397.1 SPP75128.1 CH479181 EDW31701.1 CP012528 ALC48305.1 ADTU01019952 ADTU01019953 ADTU01019954 ADTU01019955 ADTU01019956 ADTU01019957 ADTU01019958 ADTU01019959 ADTU01019960 ADTU01019961
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000283053
UP000007151
+ More
UP000235965 UP000095300 UP000005203 UP000053825 UP000215335 UP000009046 UP000008711 UP000008820 UP000092460 UP000092443 UP000091820 UP000027135 UP000078492 UP000095301 UP000183832 UP000007798 UP000008744 UP000009192 UP000007266 UP000279307 UP000002282 UP000000304 UP000037069 UP000008792 UP000268350 UP000242457 UP000001819 UP000000803 UP000192221 UP000092445 UP000078200 UP000000311 UP000008237 UP000005205 UP000069940 UP000249989 UP000092462 UP000075809 UP000076858 UP000053105 UP000092553 UP000000305
UP000235965 UP000095300 UP000005203 UP000053825 UP000215335 UP000009046 UP000008711 UP000008820 UP000092460 UP000092443 UP000091820 UP000027135 UP000078492 UP000095301 UP000183832 UP000007798 UP000008744 UP000009192 UP000007266 UP000279307 UP000002282 UP000000304 UP000037069 UP000008792 UP000268350 UP000242457 UP000001819 UP000000803 UP000192221 UP000092445 UP000078200 UP000000311 UP000008237 UP000005205 UP000069940 UP000249989 UP000092462 UP000075809 UP000076858 UP000053105 UP000092553 UP000000305
SUPFAM
SSF161070
SSF161070
ProteinModelPortal
H9JCI3
O76188
Q9U5A9
A0A194QSZ7
A0A194QH91
A0A2H1VRW7
+ More
A0A2A4JUB4 A0A2A4JNI2 A0A1E1VZ02 B2ZXL8 A0A3S2M4K9 A0A2A4JN91 A0A2A4JPM7 A0A2A4JQ43 A0A2A4JQ17 A0A2H1VSW1 A0A212EKM3 A0A1E1WV88 A0A1E1W207 A0A212EKP1 A0A2J7PKX9 A0A1I8Q6E3 A0A2J7PKT0 A0A088AJX5 A0A0L7RF06 A0A310SK53 A0A1Q3G5A3 A0A232F021 Q6VS78 E0VLW4 B3NV41 Q16YU3 A0A1B0B655 J9HFX8 Q8IST0 A0A1A9XLV3 A0A1A9WAV7 A0A067RFW9 A0A0K8TP80 U5EJQ1 A0A195DL34 A0A1I8MYH7 A0A1J1HQJ1 B4NDL8 B4GVM9 B4L7U0 D6WWF3 A0A310S8Y0 A0A3L8DNA2 B4PZQ4 B4R4T6 A0A088AJW4 A0A0L0BU43 B4MEG2 A0A3B0JYF6 A0A2A3ECG5 A0A088AJX6 Q29GB8 C7LA87 A0A2A3EB77 A0A0A1XSM6 Q9W4C5 A0A1W4V6M6 A0A336LX44 A0A1B0AE48 A0A3L8DT63 A0A1A9UT47 E2AWU5 E2BSN7 A0A158NLY9 A0A1B0B654 A0A0K8V9C6 A0A336KAS2 A0A0P6GKY2 A0A1L8DK96 A0A1L8DJX3 A0A182GFI3 A0A0P5G743 A0A336KF85 A0A1B0DCW8 A0A151XF32 A0A0P6HGT0 A0A1L8DJX4 A0A0N8A3A2 B5DZG7 A0A0M9ABU2 A0A0R3NLF5 A0A0M4E615 A0A3B0J223 A0A0P5RG87 E9G979 A0A034WJ71 A0A182GIY5 A0A0A1XHG2 A0A3B0JVN4 B4GDP4 A0A0M4EX99 A0A158NLY7
A0A2A4JUB4 A0A2A4JNI2 A0A1E1VZ02 B2ZXL8 A0A3S2M4K9 A0A2A4JN91 A0A2A4JPM7 A0A2A4JQ43 A0A2A4JQ17 A0A2H1VSW1 A0A212EKM3 A0A1E1WV88 A0A1E1W207 A0A212EKP1 A0A2J7PKX9 A0A1I8Q6E3 A0A2J7PKT0 A0A088AJX5 A0A0L7RF06 A0A310SK53 A0A1Q3G5A3 A0A232F021 Q6VS78 E0VLW4 B3NV41 Q16YU3 A0A1B0B655 J9HFX8 Q8IST0 A0A1A9XLV3 A0A1A9WAV7 A0A067RFW9 A0A0K8TP80 U5EJQ1 A0A195DL34 A0A1I8MYH7 A0A1J1HQJ1 B4NDL8 B4GVM9 B4L7U0 D6WWF3 A0A310S8Y0 A0A3L8DNA2 B4PZQ4 B4R4T6 A0A088AJW4 A0A0L0BU43 B4MEG2 A0A3B0JYF6 A0A2A3ECG5 A0A088AJX6 Q29GB8 C7LA87 A0A2A3EB77 A0A0A1XSM6 Q9W4C5 A0A1W4V6M6 A0A336LX44 A0A1B0AE48 A0A3L8DT63 A0A1A9UT47 E2AWU5 E2BSN7 A0A158NLY9 A0A1B0B654 A0A0K8V9C6 A0A336KAS2 A0A0P6GKY2 A0A1L8DK96 A0A1L8DJX3 A0A182GFI3 A0A0P5G743 A0A336KF85 A0A1B0DCW8 A0A151XF32 A0A0P6HGT0 A0A1L8DJX4 A0A0N8A3A2 B5DZG7 A0A0M9ABU2 A0A0R3NLF5 A0A0M4E615 A0A3B0J223 A0A0P5RG87 E9G979 A0A034WJ71 A0A182GIY5 A0A0A1XHG2 A0A3B0JVN4 B4GDP4 A0A0M4EX99 A0A158NLY7
PDB
4XPT
E-value=2.46203e-90,
Score=849
Ontologies
GO
PANTHER
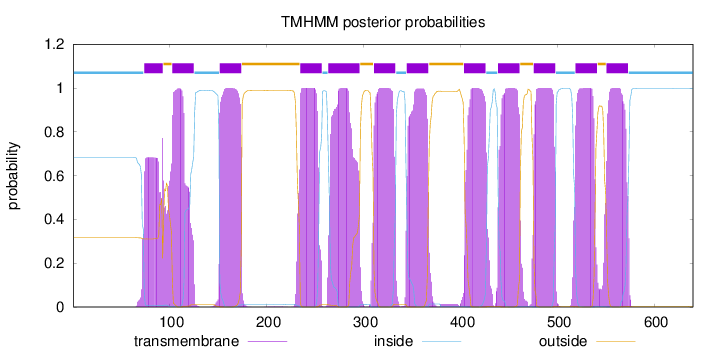
Topology
Subcellular location
Membrane
Length:
640
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
266.98648
Exp number, first 60 AAs:
0
Total prob of N-in:
0.68319
inside
1 - 73
TMhelix
74 - 93
outside
94 - 102
TMhelix
103 - 125
inside
126 - 151
TMhelix
152 - 174
outside
175 - 234
TMhelix
235 - 257
inside
258 - 263
TMhelix
264 - 296
outside
297 - 310
TMhelix
311 - 333
inside
334 - 344
TMhelix
345 - 367
outside
368 - 403
TMhelix
404 - 426
inside
427 - 438
TMhelix
439 - 461
outside
462 - 475
TMhelix
476 - 498
inside
499 - 518
TMhelix
519 - 541
outside
542 - 550
TMhelix
551 - 573
inside
574 - 640
Population Genetic Test Statistics
Pi
208.413179
Theta
174.320169
Tajima's D
0.689516
CLR
0.073049
CSRT
0.565871706414679
Interpretation
Uncertain
