Gene
KWMTBOMO10332
Pre Gene Modal
BGIBMGA007232
Annotation
PREDICTED:_diacylglycerol_kinase_epsilon_isoform_X2_[Amyelois_transitella]
Full name
Diacylglycerol kinase
Location in the cell
PlasmaMembrane Reliability : 1.519
Sequence
CDS
ATGTTGGCTGTAGTTGGATTGTTTTGTGAGTGTTGTGCTGTTAGTGCATGCAAGAAATGCTACAAGACAGTGGATAAGAGATATAAATGTAAACAATTAACTTGGCCTTCTAATAAAATATTTCCTCATCAGTGGGTTGATGTTGGAATTTCAAGAACAGACATTGCTGAAAATAATGATCATAGTGATGATAGTACTAAGAAATATTTTTGTAGCTGGTGCCAAAGAATAAAGTTGTGTAATAATACTAGGGATTTGAAAACTGAGCAATGTGACTTCCATAAATATAGCAATATTATCATTCCACCCACATCAGTACAAATGGAGAAAGGCAATGTTATTAGTATTCAGCCGGTTAATAACAGCAATTGGGAACCTCTGATCATTATTGCAAACAGGAAATCCGGCAGCAACAGGAGTGATGAGATACTTTCATTATTTAGAGGATTACTCAATCCAATTCAGGTGATTGACGTAAGCGTGACCTCCCCTGAAAGTGTGGTGCGGTGGCTCCCTGAGAAATGTCGTGTGTTGGTCGCAGGTGGGGACGGCACCGTTGCTTGGGTACTTAATGCTTTATTCGCTGCGCCGCACATCAAGGCGGCCGTGGCTATCTTCCCTATGGGGACCGGTAATGATCTGTCTCGTGTGCTCGGCTGGGGTCCGGGCTGTCCGTCATACATTGATGCACATTCAATCATTAGCAGCATAAAACAAGCAACAGTTGAACAATTGGACAGATGGAAAATTATTATTAAACCAAAGCGTGGCCGTCTCGGTCGGCGGGCACCGGAGCGTATATTGCACGCTTACAACTATGCGAGTATTGGAGTAGACGCTCAGGTCGCGCTCGACTTCCATCGCGCCAGGACGCAGTTCTTATATCGATACGCAAGCAGGACCCTTAATTACATAGCGTACGTGTTTCTGGGCGTGGGTCGTGCGCTGGACGACGGCGGGTGCGAGGGGCTGGAGCGGCGGGTGCGGTGCTGCGCGGATGGCGTGGCGTTGACCCTGCCGCCGCTGCAGGCTCTGCTCGCACTCAACATACCCTCCTGGGGCGCAGGCGTGCACCTCTGGAGTATGGGAAGTGAAGATGAAGTCGAAGAACAAAGCATGAGCGATGGAAAATTGGAGGTGGTCGGCATTTCGTCTTCATTTCACATTGCAAGACTTCAGTGCGGTCTGGCTGAACCTTATCGTTTCACACAAGCGTCCAATCTCAGAATTGATCTAGAAGGTTCGTGTGCGATGCAAGTGGACGGCGAGCCCTGGATGCAGGGTCCGGCCACTATTTTAGTGGAGCCGGCTGGACAAAGCACCATGCTGAGAGCGAACATAAAAAGATCTGAATGTTAA
Protein
MLAVVGLFCECCAVSACKKCYKTVDKRYKCKQLTWPSNKIFPHQWVDVGISRTDIAENNDHSDDSTKKYFCSWCQRIKLCNNTRDLKTEQCDFHKYSNIIIPPTSVQMEKGNVISIQPVNNSNWEPLIIIANRKSGSNRSDEILSLFRGLLNPIQVIDVSVTSPESVVRWLPEKCRVLVAGGDGTVAWVLNALFAAPHIKAAVAIFPMGTGNDLSRVLGWGPGCPSYIDAHSIISSIKQATVEQLDRWKIIIKPKRGRLGRRAPERILHAYNYASIGVDAQVALDFHRARTQFLYRYASRTLNYIAYVFLGVGRALDDGGCEGLERRVRCCADGVALTLPPLQALLALNIPSWGAGVHLWSMGSEDEVEEQSMSDGKLEVVGISSSFHIARLQCGLAEPYRFTQASNLRIDLEGSCAMQVDGEPWMQGPATILVEPAGQSTMLRANIKRSEC
Summary
Catalytic Activity
a 1,2-diacyl-sn-glycerol + ATP = a 1,2-diacyl-sn-glycero-3-phosphate + ADP + H(+)
Similarity
Belongs to the eukaryotic diacylglycerol kinase family.
Feature
chain Diacylglycerol kinase
Uniprot
A0A2H1V771
A0A2A4JRX5
A0A3S2LUW9
A0A194QH30
A0A194QX86
A0A212EIE2
+ More
E0VQ75 A0A0L7QK09 A0A195DB38 A0A195AX69 A0A158NLP5 A0A2J7PXH5 K7IWS4 A0A151X811 A0A195C1Y5 A0A232F6G1 F4WXH5 A0A026WJ93 A0A3L8E2R7 A0A1B6C2W1 A0A067QNK5 A0A0T6AYK2 A0A195F593 W5JS06 A0A182FI18 A0A154P9Q0 A0A1B0CE52 A0A1I8NPY8 A0A2P8Y5R9 B3NRV5 A0A0K8VXW9 A0A3B0J0D0 A0A224XMK3 B4P4Y9 A0A1W4VPT5 A0A034WSQ1 A0A1I8N1X9 B3MG14 A0A139WDJ5 A0A0A1WG30 B4KQ02 W8BTH7 B4J883 A0A1A9V247 A0A146M925 F3YD90 B4MDU6 B4HPP2 Q7K579 A0A182WI69 A0A182JDF5 A0A182QQL5 O44334 Q28X81 B4H547 A0A1B0AB78 B4QDG9 A0A1A9X4S0 A0A182PEY2 A0A182SNN6 A0A182K9Y2 E2B3W8 A0A1A9XPJ6 A0A1B0BXA7 A0A182LXJ6 B4N5Z8 A0A182N7J3 A0A0L0C393 A0A2A3E352 A0A182YJI6 Q7PT01 A0A0P4VSY5 A0A0M4EI43 A0A0N0U7D2 E2AH29 A0A171AH78 A0A182R9A4 A0A224YRE2 A0A131YKC7 A0A2R5LDC1 A0A1S4FU05 A0A310SX15 A0A0P5LDM1 A0A182GS87 A0A0N8BG93 E9G1Y7 A0A084WLA8 A0A0P5DYW8 A0A1B6EKU2 A0A0P5W951 A0A1B0D780 A0A0P6IVW4 A0A0K8V5U8 A0A0P5WTH3 B0WPH8 A0A1Q3FDA3 A0A0P4Z6X5 A0A1Y1MA97 A0A131XTD3 Q16PA7
E0VQ75 A0A0L7QK09 A0A195DB38 A0A195AX69 A0A158NLP5 A0A2J7PXH5 K7IWS4 A0A151X811 A0A195C1Y5 A0A232F6G1 F4WXH5 A0A026WJ93 A0A3L8E2R7 A0A1B6C2W1 A0A067QNK5 A0A0T6AYK2 A0A195F593 W5JS06 A0A182FI18 A0A154P9Q0 A0A1B0CE52 A0A1I8NPY8 A0A2P8Y5R9 B3NRV5 A0A0K8VXW9 A0A3B0J0D0 A0A224XMK3 B4P4Y9 A0A1W4VPT5 A0A034WSQ1 A0A1I8N1X9 B3MG14 A0A139WDJ5 A0A0A1WG30 B4KQ02 W8BTH7 B4J883 A0A1A9V247 A0A146M925 F3YD90 B4MDU6 B4HPP2 Q7K579 A0A182WI69 A0A182JDF5 A0A182QQL5 O44334 Q28X81 B4H547 A0A1B0AB78 B4QDG9 A0A1A9X4S0 A0A182PEY2 A0A182SNN6 A0A182K9Y2 E2B3W8 A0A1A9XPJ6 A0A1B0BXA7 A0A182LXJ6 B4N5Z8 A0A182N7J3 A0A0L0C393 A0A2A3E352 A0A182YJI6 Q7PT01 A0A0P4VSY5 A0A0M4EI43 A0A0N0U7D2 E2AH29 A0A171AH78 A0A182R9A4 A0A224YRE2 A0A131YKC7 A0A2R5LDC1 A0A1S4FU05 A0A310SX15 A0A0P5LDM1 A0A182GS87 A0A0N8BG93 E9G1Y7 A0A084WLA8 A0A0P5DYW8 A0A1B6EKU2 A0A0P5W951 A0A1B0D780 A0A0P6IVW4 A0A0K8V5U8 A0A0P5WTH3 B0WPH8 A0A1Q3FDA3 A0A0P4Z6X5 A0A1Y1MA97 A0A131XTD3 Q16PA7
EC Number
2.7.1.107
Pubmed
26354079
22118469
20566863
21347285
20075255
28648823
+ More
21719571 24508170 30249741 24845553 20920257 23761445 29403074 17994087 17550304 25348373 25315136 18362917 19820115 25830018 24495485 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 11102369 15632085 22936249 20798317 26108605 25244985 12364791 27129103 28797301 26830274 17510324 26483478 21292972 24438588 28004739
21719571 24508170 30249741 24845553 20920257 23761445 29403074 17994087 17550304 25348373 25315136 18362917 19820115 25830018 24495485 26823975 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 11102369 15632085 22936249 20798317 26108605 25244985 12364791 27129103 28797301 26830274 17510324 26483478 21292972 24438588 28004739
EMBL
ODYU01000787
SOQ36104.1
NWSH01000726
PCG74569.1
RSAL01000198
RVE44539.1
+ More
KQ459232 KPJ02751.1 KQ461155 KPJ08186.1 AGBW02014664 OWR41256.1 DS235392 EEB15531.1 KQ414989 KOC58977.1 KQ981042 KYN10103.1 KQ976716 KYM76843.1 ADTU01002188 NEVH01020858 PNF21026.1 AAZX01004127 KQ982431 KYQ56503.1 KQ978344 KYM94857.1 NNAY01000895 OXU26029.1 GL888424 EGI61141.1 KK107218 EZA55164.1 QOIP01000001 RLU26489.1 GEDC01029699 JAS07599.1 KK853134 KDR10864.1 LJIG01022489 KRT80304.1 KQ981805 KYN35628.1 ADMH02000632 ETN65524.1 KQ434850 KZC08572.1 AJWK01008581 AJWK01008582 PYGN01000896 PSN39597.1 CH954179 EDV56257.1 GDHF01008879 JAI43435.1 OUUW01000001 SPP74057.1 GFTR01006604 JAW09822.1 CM000158 EDW90710.1 GAKP01000351 JAC58601.1 CH902619 EDV36709.1 KQ971361 KYB25925.1 GBXI01016949 JAC97342.1 CH933808 EDW08104.1 GAMC01010004 JAB96551.1 CH916367 EDW01220.1 GDHC01002864 JAQ15765.1 BT126251 AEB39612.1 CH940662 EDW58711.1 CH480816 EDW47626.1 AE013599 AY050241 AAF58458.1 AAK84940.1 AXCN02000576 AF017783 AAB97514.1 CM000071 EAL26435.2 CH479210 EDW32883.1 CM000362 CM002911 EDX06834.1 KMY93336.1 GL445407 EFN89637.1 JXJN01022149 AXCM01006814 CH964154 EDW79787.1 JRES01000960 KNC26736.1 KZ288451 PBC25569.1 AAAB01008807 EAA04759.5 GDKW01001506 JAI55089.1 CP012524 ALC40692.1 KQ435711 KOX79421.1 GL439470 EFN67260.1 GEMB01001070 JAS02077.1 GFPF01007183 MAA18329.1 GEDV01010196 JAP78361.1 GGLE01003343 MBY07469.1 KQ759840 OAD62646.1 GDIQ01181713 JAK70012.1 JXUM01016839 KQ560467 KXJ82264.1 GDIQ01180715 JAK71010.1 GL732529 EFX86572.1 ATLV01024210 KE525350 KFB51002.1 GDIP01148944 LRGB01003146 JAJ74458.1 KZS04014.1 GECZ01031215 JAS38554.1 GDIP01102692 JAM01023.1 AJVK01026697 GDIQ01027056 JAN67681.1 GDHF01028579 GDHF01018181 JAI23735.1 JAI34133.1 GDIP01082338 JAM21377.1 DS232024 EDS32366.1 GFDL01009532 JAV25513.1 GDIP01218584 JAJ04818.1 GEZM01036508 JAV82614.1 GEFM01005517 JAP70279.1 CH477789 EAT36194.1
KQ459232 KPJ02751.1 KQ461155 KPJ08186.1 AGBW02014664 OWR41256.1 DS235392 EEB15531.1 KQ414989 KOC58977.1 KQ981042 KYN10103.1 KQ976716 KYM76843.1 ADTU01002188 NEVH01020858 PNF21026.1 AAZX01004127 KQ982431 KYQ56503.1 KQ978344 KYM94857.1 NNAY01000895 OXU26029.1 GL888424 EGI61141.1 KK107218 EZA55164.1 QOIP01000001 RLU26489.1 GEDC01029699 JAS07599.1 KK853134 KDR10864.1 LJIG01022489 KRT80304.1 KQ981805 KYN35628.1 ADMH02000632 ETN65524.1 KQ434850 KZC08572.1 AJWK01008581 AJWK01008582 PYGN01000896 PSN39597.1 CH954179 EDV56257.1 GDHF01008879 JAI43435.1 OUUW01000001 SPP74057.1 GFTR01006604 JAW09822.1 CM000158 EDW90710.1 GAKP01000351 JAC58601.1 CH902619 EDV36709.1 KQ971361 KYB25925.1 GBXI01016949 JAC97342.1 CH933808 EDW08104.1 GAMC01010004 JAB96551.1 CH916367 EDW01220.1 GDHC01002864 JAQ15765.1 BT126251 AEB39612.1 CH940662 EDW58711.1 CH480816 EDW47626.1 AE013599 AY050241 AAF58458.1 AAK84940.1 AXCN02000576 AF017783 AAB97514.1 CM000071 EAL26435.2 CH479210 EDW32883.1 CM000362 CM002911 EDX06834.1 KMY93336.1 GL445407 EFN89637.1 JXJN01022149 AXCM01006814 CH964154 EDW79787.1 JRES01000960 KNC26736.1 KZ288451 PBC25569.1 AAAB01008807 EAA04759.5 GDKW01001506 JAI55089.1 CP012524 ALC40692.1 KQ435711 KOX79421.1 GL439470 EFN67260.1 GEMB01001070 JAS02077.1 GFPF01007183 MAA18329.1 GEDV01010196 JAP78361.1 GGLE01003343 MBY07469.1 KQ759840 OAD62646.1 GDIQ01181713 JAK70012.1 JXUM01016839 KQ560467 KXJ82264.1 GDIQ01180715 JAK71010.1 GL732529 EFX86572.1 ATLV01024210 KE525350 KFB51002.1 GDIP01148944 LRGB01003146 JAJ74458.1 KZS04014.1 GECZ01031215 JAS38554.1 GDIP01102692 JAM01023.1 AJVK01026697 GDIQ01027056 JAN67681.1 GDHF01028579 GDHF01018181 JAI23735.1 JAI34133.1 GDIP01082338 JAM21377.1 DS232024 EDS32366.1 GFDL01009532 JAV25513.1 GDIP01218584 JAJ04818.1 GEZM01036508 JAV82614.1 GEFM01005517 JAP70279.1 CH477789 EAT36194.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000053240
UP000007151
UP000009046
+ More
UP000053825 UP000078492 UP000078540 UP000005205 UP000235965 UP000002358 UP000075809 UP000078542 UP000215335 UP000007755 UP000053097 UP000279307 UP000027135 UP000078541 UP000000673 UP000069272 UP000076502 UP000092461 UP000095300 UP000245037 UP000008711 UP000268350 UP000002282 UP000192221 UP000095301 UP000007801 UP000007266 UP000009192 UP000001070 UP000078200 UP000008792 UP000001292 UP000000803 UP000075920 UP000075880 UP000075886 UP000001819 UP000008744 UP000092445 UP000000304 UP000091820 UP000075885 UP000075901 UP000075881 UP000008237 UP000092443 UP000092460 UP000075883 UP000007798 UP000075884 UP000037069 UP000242457 UP000076408 UP000007062 UP000092553 UP000053105 UP000000311 UP000075900 UP000069940 UP000249989 UP000000305 UP000030765 UP000076858 UP000092462 UP000002320 UP000008820
UP000053825 UP000078492 UP000078540 UP000005205 UP000235965 UP000002358 UP000075809 UP000078542 UP000215335 UP000007755 UP000053097 UP000279307 UP000027135 UP000078541 UP000000673 UP000069272 UP000076502 UP000092461 UP000095300 UP000245037 UP000008711 UP000268350 UP000002282 UP000192221 UP000095301 UP000007801 UP000007266 UP000009192 UP000001070 UP000078200 UP000008792 UP000001292 UP000000803 UP000075920 UP000075880 UP000075886 UP000001819 UP000008744 UP000092445 UP000000304 UP000091820 UP000075885 UP000075901 UP000075881 UP000008237 UP000092443 UP000092460 UP000075883 UP000007798 UP000075884 UP000037069 UP000242457 UP000076408 UP000007062 UP000092553 UP000053105 UP000000311 UP000075900 UP000069940 UP000249989 UP000000305 UP000030765 UP000076858 UP000092462 UP000002320 UP000008820
Interpro
IPR002110
Ankyrin_rpt
+ More
IPR037607 DGK
IPR016064 NAD/diacylglycerol_kinase_sf
IPR017438 ATP-NAD_kinase_N
IPR001206 Diacylglycerol_kinase_cat_dom
IPR000756 Diacylglycerol_kin_accessory
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR002219 PE/DAG-bd
IPR000719 Prot_kinase_dom
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR037607 DGK
IPR016064 NAD/diacylglycerol_kinase_sf
IPR017438 ATP-NAD_kinase_N
IPR001206 Diacylglycerol_kinase_cat_dom
IPR000756 Diacylglycerol_kin_accessory
IPR020683 Ankyrin_rpt-contain_dom
IPR036770 Ankyrin_rpt-contain_sf
IPR002219 PE/DAG-bd
IPR000719 Prot_kinase_dom
IPR008271 Ser/Thr_kinase_AS
IPR011009 Kinase-like_dom_sf
Gene 3D
ProteinModelPortal
A0A2H1V771
A0A2A4JRX5
A0A3S2LUW9
A0A194QH30
A0A194QX86
A0A212EIE2
+ More
E0VQ75 A0A0L7QK09 A0A195DB38 A0A195AX69 A0A158NLP5 A0A2J7PXH5 K7IWS4 A0A151X811 A0A195C1Y5 A0A232F6G1 F4WXH5 A0A026WJ93 A0A3L8E2R7 A0A1B6C2W1 A0A067QNK5 A0A0T6AYK2 A0A195F593 W5JS06 A0A182FI18 A0A154P9Q0 A0A1B0CE52 A0A1I8NPY8 A0A2P8Y5R9 B3NRV5 A0A0K8VXW9 A0A3B0J0D0 A0A224XMK3 B4P4Y9 A0A1W4VPT5 A0A034WSQ1 A0A1I8N1X9 B3MG14 A0A139WDJ5 A0A0A1WG30 B4KQ02 W8BTH7 B4J883 A0A1A9V247 A0A146M925 F3YD90 B4MDU6 B4HPP2 Q7K579 A0A182WI69 A0A182JDF5 A0A182QQL5 O44334 Q28X81 B4H547 A0A1B0AB78 B4QDG9 A0A1A9X4S0 A0A182PEY2 A0A182SNN6 A0A182K9Y2 E2B3W8 A0A1A9XPJ6 A0A1B0BXA7 A0A182LXJ6 B4N5Z8 A0A182N7J3 A0A0L0C393 A0A2A3E352 A0A182YJI6 Q7PT01 A0A0P4VSY5 A0A0M4EI43 A0A0N0U7D2 E2AH29 A0A171AH78 A0A182R9A4 A0A224YRE2 A0A131YKC7 A0A2R5LDC1 A0A1S4FU05 A0A310SX15 A0A0P5LDM1 A0A182GS87 A0A0N8BG93 E9G1Y7 A0A084WLA8 A0A0P5DYW8 A0A1B6EKU2 A0A0P5W951 A0A1B0D780 A0A0P6IVW4 A0A0K8V5U8 A0A0P5WTH3 B0WPH8 A0A1Q3FDA3 A0A0P4Z6X5 A0A1Y1MA97 A0A131XTD3 Q16PA7
E0VQ75 A0A0L7QK09 A0A195DB38 A0A195AX69 A0A158NLP5 A0A2J7PXH5 K7IWS4 A0A151X811 A0A195C1Y5 A0A232F6G1 F4WXH5 A0A026WJ93 A0A3L8E2R7 A0A1B6C2W1 A0A067QNK5 A0A0T6AYK2 A0A195F593 W5JS06 A0A182FI18 A0A154P9Q0 A0A1B0CE52 A0A1I8NPY8 A0A2P8Y5R9 B3NRV5 A0A0K8VXW9 A0A3B0J0D0 A0A224XMK3 B4P4Y9 A0A1W4VPT5 A0A034WSQ1 A0A1I8N1X9 B3MG14 A0A139WDJ5 A0A0A1WG30 B4KQ02 W8BTH7 B4J883 A0A1A9V247 A0A146M925 F3YD90 B4MDU6 B4HPP2 Q7K579 A0A182WI69 A0A182JDF5 A0A182QQL5 O44334 Q28X81 B4H547 A0A1B0AB78 B4QDG9 A0A1A9X4S0 A0A182PEY2 A0A182SNN6 A0A182K9Y2 E2B3W8 A0A1A9XPJ6 A0A1B0BXA7 A0A182LXJ6 B4N5Z8 A0A182N7J3 A0A0L0C393 A0A2A3E352 A0A182YJI6 Q7PT01 A0A0P4VSY5 A0A0M4EI43 A0A0N0U7D2 E2AH29 A0A171AH78 A0A182R9A4 A0A224YRE2 A0A131YKC7 A0A2R5LDC1 A0A1S4FU05 A0A310SX15 A0A0P5LDM1 A0A182GS87 A0A0N8BG93 E9G1Y7 A0A084WLA8 A0A0P5DYW8 A0A1B6EKU2 A0A0P5W951 A0A1B0D780 A0A0P6IVW4 A0A0K8V5U8 A0A0P5WTH3 B0WPH8 A0A1Q3FDA3 A0A0P4Z6X5 A0A1Y1MA97 A0A131XTD3 Q16PA7
PDB
4WRR
E-value=0.00857556,
Score=93
Ontologies
KEGG
PATHWAY
GO
PANTHER
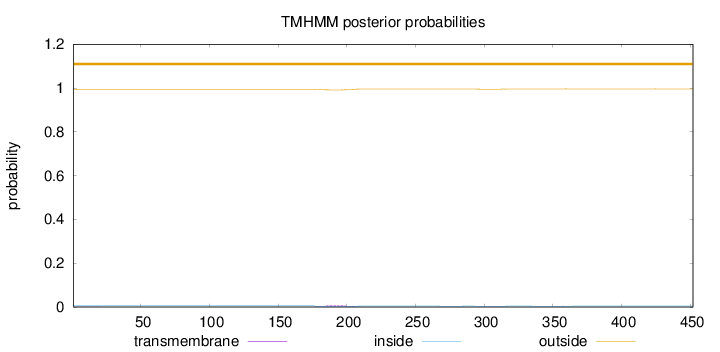
Topology
Length:
452
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.24136
Exp number, first 60 AAs:
0.00236
Total prob of N-in:
0.00577
outside
1 - 452
Population Genetic Test Statistics
Pi
193.184692
Theta
151.688524
Tajima's D
0.908765
CLR
0.165014
CSRT
0.634768261586921
Interpretation
Uncertain
