Gene
KWMTBOMO10328
Pre Gene Modal
BGIBMGA007240
Annotation
PREDICTED:_transducin_beta-like_protein_3_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.64
Sequence
CDS
ATGTCAAATTTAAAAGAAATATATGAGAAAAGTGCAGAGTTTACTGCATTTTATACAGGTGGTGATATACAGTGGACAAATGATGGCTTACAAATGCTTTGCCTATGTGATGAAGCTATTAAGGTGGTTGAAGTCGACACTCTAATCACTTCTCTTATTATTGGCGAAACTGAGGAAGATGCAGAAAATGACCAGATTTATACATTTAAAATATCACATAGTAATCAAAGTATTGTTACTGCACACAAAAGTGGCCTAATTAAAATGTGGGACAAAGAAACTGGAGTCCAACTAAAAATTTGGAGATCAAGCCATAAAGGACCTGTTGTGAAGTTAGCCTTTGATTCAACAGATGAAAATATTGCTTCAGGAGGTTCCGACGGTAATGTGAGACTGTGGGATGTAGGACATAATACCTGTACTAGCAGTCTTCGTGGAGCTATGGGTGTTTTCTCTGTGTTAGAGTATCATCCAGATTCTTCTAAACAATTAGTATTTGGAGCAGCAGATGATACAAAGATTAGATCCTGGTGTTCTAAAACTGGTAAAGAGCAATTAGTATATTCAGGTCATTTTAGTAAGGTAACTGCTTTGCTATTCACACTTGATGGTAAATACATGGTCAGCTCTGGGAGGGACAGAGTACTCATATTGTGGGATTTAAATGTTAGCAAAGCTCTTAAAGTTGTTCCTGTGTATGAAGGAATTGAATCCTGCATTCTCTTACCACCTTCATTTAAAATCCCAAATTTTAATAAAAAGCTGGAAACTGAAGGGGTTTATGTTGCTTGTGCTGGAGAAAAAGGAATTGTTAAAGTATGGAATTTGTCAATGAGCCGACTCATGTTTGAACAAATCAATAGTTTAGTATCCCCGTCAACTGAAGAAGGAGGGCTGTCAATAACGCACCTACTATATAATCAAGCAAGAAATATGGTCTCTGTAGTGACAGTTGATCATAACATTATAATACATGACCTAGAAACATTTGACTCTGTGAAACAGATGATTGGTTTCACTGATGAAGTTCTTGACATAATATTTTTGGGAAAAAACGAGAACCATATAATTGTTGCCACAAATAGCCAAGATCTGAAGTATTATGATTTAGAAACTATGAATTGTCAAATTGTTAAAGGTCACACAGATATAGTGCTCTCCTTAGCAAACTTTCCAACTAAACTTGAAATGTTTGTTTCATCAAGTAAAGACAATAGCGTTCGATTGTGGCTTCAAACACCAAACAACACTATTCATTGCCTTGCAGTTGGAACTCGCCATACTGCTTCTGTTGGGTCTGTTTACACTTCACAAACATCCGATGATTTCTTTACCTCGGTTAGTCAAGATAATTGCATAAAAGTTTGGACTGTATGTAAGCCTGCAGAAAAATGTGTAGAACAAAATCTTAAATGTAGTCACACTGAATTAGCTCACCAAATGGACATTAATTGTGTTGCAGTATCACCCAATGATAAAATGATAGCTACTGGTTCACAAGATAAAACTGCAAAGATTTGGGCAGAAGACTTGACACTGTTAGGTGTATTAAAAGGTCACAGAAGGGGAGTGTGGTGTGTAAGATTTTCTCCTGTCGATCAAGTAGTGCTAACATCTTCAGCTGACAGTTCCATTAAATTATGGTCCATAAGTGATTTAAGTTGCCTAAAAACTTTCGAAGGTCATGAAAGTTCTGTTCTGAAAGTTGAGTTCATTAGTAAAGGCCAACAGATTTTATCCAGTGGAGCTGATGGATTATTAAAGTTATGGACTATCAAAACATCAGAAAGTAAAATGTCATTAGACAACCATGATGGTAAAGTGTGGTCATTAGCTATATCCAAAAATGAATCATACATAATCACAGGTGGTTCTGATTCAAAATTAGTGAAATTAAAAGATGTAACATTGGAACAAAGAGAAAGAAGGACCAAGGAGAGAGAAGCACTGATTTTGCAAGAACAGGAATTAAATAACTTACTTCATGACAAAAAATTACTCAAAGCTTTAAAATTAGCATTACGAATGGACAGGCCACTGCACGTGTTAAAAATAACCGCTCGCGATTTCGCGCCACTGCGCGGATTATATGTTTATTTCATCATGATTAAAAAAATCACAATCTATGCCCTGCTGACGGTTTTCATTGAAAATTCTCAAGGACAAAAATCGCAACAGATAGCGCCCTTAGAATGTCCAAGTTTAGACAGCGGAGACAGTTTTACTCAGTCGGAGTTCTTATCAAGACTCGCAAACGAATGCCGCTACGACCGGCTACTGCTGCCTACCTATCAGACCGGTGACGTCGTCTACGTCCATGCCAGCGCCTACGTTTATTTCATTCAACCAGCAGAAGCACACGATTTGAATTTTAAACTTCACTTCCTTCTGCAACTACGATGGACGGACCCGCGCCTAGCGTACACACTCTACTCTCCGAAAAGATCTAAAATCATAGGTGAAAATGATCTGAAACAAAGAATATGGGTGCCACATCTATACATGTCGAACGAACAGTCGTCGAGTCTCATGGGAACCGACAGTAAAGACGTTCTCATTTCAATCGCTCCCGACGGTGAAGTGCTTTTCAGCAGGCGAATGCAGGCAGTGTTGTACTGTTGGATGAATCTACAGAAATTCCCCTTTGACGATCAGACATGCTCTATGAACTTGGAAAGCTGGAAATATAATGCATCGATTCTACGTTTAATGTGGGAAAAGGATAATCCAGTTCGCTTATCATCGGAACTACACCTGACAGAATATTCTCTCATGGATTTTTGGACGAACGAATCTGTTGTCAGAGGCGATATAGCCAACATGCGTCTTGGTGGAGCGGGTAATTATAGCGCCTTAAAATTCACATTCAAACTTGGTCGCGAGGTCGGTTACTATCTCATGGATTATTTTATTCCATCGATGATGATAGTGGCAATGTCGTGGGTTACATTTTGGCTACAAGCGGACGCATCAGCACCAAGAATAACTTTAGGTACAAGCACTATGTTATCGTTTATAACACTGGCATCATCGCAAGCGAAGACACTACCTAAAGTTTCATATATCAAAGCTAGCGAGGTCTGGTTCTTAGGCTGCACCGGTTTCATCTTCTCAGCTCTTGTAGAATTTGCTTTTGTCAACACCATTTGGAGAAGAAAGAAAGCGGTAAACTTAAAAAAGGTGAATAGCAAATACATTTTGAAGAGCACGCTTACTCCGCGTTTGGCCCGTAAAGAACTTCAAAAAGAAATACAAGAATCTTCACCGCAACTTTCGAAATCGAGATCTTGCTCGTCGCTGCAGCAGGAGGGCAGCTGCAACGACCCAGCCGGTCCCGGTTACAACAACTACCTCACTGTGCACCCGAACAGGGAATTGTAA
Protein
MSNLKEIYEKSAEFTAFYTGGDIQWTNDGLQMLCLCDEAIKVVEVDTLITSLIIGETEEDAENDQIYTFKISHSNQSIVTAHKSGLIKMWDKETGVQLKIWRSSHKGPVVKLAFDSTDENIASGGSDGNVRLWDVGHNTCTSSLRGAMGVFSVLEYHPDSSKQLVFGAADDTKIRSWCSKTGKEQLVYSGHFSKVTALLFTLDGKYMVSSGRDRVLILWDLNVSKALKVVPVYEGIESCILLPPSFKIPNFNKKLETEGVYVACAGEKGIVKVWNLSMSRLMFEQINSLVSPSTEEGGLSITHLLYNQARNMVSVVTVDHNIIIHDLETFDSVKQMIGFTDEVLDIIFLGKNENHIIVATNSQDLKYYDLETMNCQIVKGHTDIVLSLANFPTKLEMFVSSSKDNSVRLWLQTPNNTIHCLAVGTRHTASVGSVYTSQTSDDFFTSVSQDNCIKVWTVCKPAEKCVEQNLKCSHTELAHQMDINCVAVSPNDKMIATGSQDKTAKIWAEDLTLLGVLKGHRRGVWCVRFSPVDQVVLTSSADSSIKLWSISDLSCLKTFEGHESSVLKVEFISKGQQILSSGADGLLKLWTIKTSESKMSLDNHDGKVWSLAISKNESYIITGGSDSKLVKLKDVTLEQRERRTKEREALILQEQELNNLLHDKKLLKALKLALRMDRPLHVLKITARDFAPLRGLYVYFIMIKKITIYALLTVFIENSQGQKSQQIAPLECPSLDSGDSFTQSEFLSRLANECRYDRLLLPTYQTGDVVYVHASAYVYFIQPAEAHDLNFKLHFLLQLRWTDPRLAYTLYSPKRSKIIGENDLKQRIWVPHLYMSNEQSSSLMGTDSKDVLISIAPDGEVLFSRRMQAVLYCWMNLQKFPFDDQTCSMNLESWKYNASILRLMWEKDNPVRLSSELHLTEYSLMDFWTNESVVRGDIANMRLGGAGNYSALKFTFKLGREVGYYLMDYFIPSMMIVAMSWVTFWLQADASAPRITLGTSTMLSFITLASSQAKTLPKVSYIKASEVWFLGCTGFIFSALVEFAFVNTIWRRKKAVNLKKVNSKYILKSTLTPRLARKELQKEIQESSPQLSKSRSCSSLQQEGSCNDPAGPGYNNYLTVHPNREL
Summary
Similarity
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Uniprot
EMBL
Proteomes
Interpro
IPR036322
WD40_repeat_dom_sf
+ More
IPR019775 WD40_repeat_CS
IPR018000 Neurotransmitter_ion_chnl_CS
IPR020472 G-protein_beta_WD-40_rep
IPR006202 Neur_chan_lig-bd
IPR006028 GABAA/Glycine_rcpt
IPR001680 WD40_repeat
IPR006029 Neurotrans-gated_channel_TM
IPR036734 Neur_chan_lig-bd_sf
IPR017986 WD40_repeat_dom
IPR036719 Neuro-gated_channel_TM_sf
IPR013934 SSU_processome_Utp13
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR006201 Neur_channel
IPR019775 WD40_repeat_CS
IPR018000 Neurotransmitter_ion_chnl_CS
IPR020472 G-protein_beta_WD-40_rep
IPR006202 Neur_chan_lig-bd
IPR006028 GABAA/Glycine_rcpt
IPR001680 WD40_repeat
IPR006029 Neurotrans-gated_channel_TM
IPR036734 Neur_chan_lig-bd_sf
IPR017986 WD40_repeat_dom
IPR036719 Neuro-gated_channel_TM_sf
IPR013934 SSU_processome_Utp13
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR006201 Neur_channel
Gene 3D
ProteinModelPortal
PDB
5OQL
E-value=2.6414e-63,
Score=618
Ontologies
GO
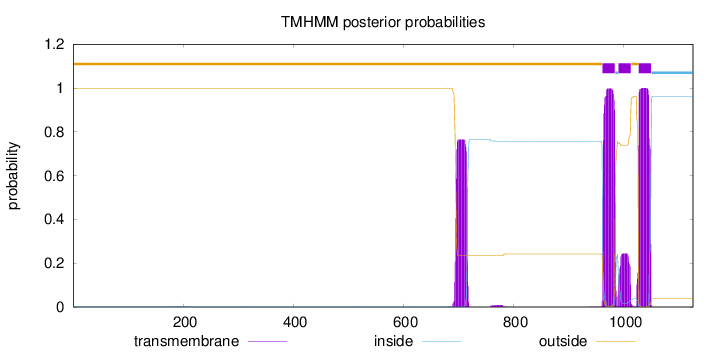
Topology
Length:
1126
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
66.98941
Exp number, first 60 AAs:
0.00054
Total prob of N-in:
0.00086
outside
1 - 961
TMhelix
962 - 984
inside
985 - 990
TMhelix
991 - 1013
outside
1014 - 1027
TMhelix
1028 - 1050
inside
1051 - 1126
Population Genetic Test Statistics
Pi
186.843082
Theta
179.676692
Tajima's D
-0.782112
CLR
101.069309
CSRT
0.179291035448228
Interpretation
Uncertain
