Pre Gene Modal
BGIBMGA007222
Annotation
RPA43_N_protein_[Bombyx_mori]
Full name
DNA-directed RNA polymerase I subunit RPA43
Alternative Name
Twist neighbor protein
Location in the cell
Cytoplasmic Reliability : 2.528
Sequence
CDS
ATGTCTAAAATCATAAAATTTGACCTACGAGAGTTAAAACAGTTGGCAAACGACAAAAATTCTTGTTTGGTTGTAAAAAAGATTACACAAAATCTGGCTTTACAGCCATGGTGCCTTGGAAATATTAAAGAATCTATAACAAACTTATTAGATTATAAAGTTGGTAAATTTGATAAAGAGTTGAACGGTGTCTTAGTAAGCTATAAAAATCCTTGTATACTTCAGAATGTTGGCACTATACGCAATGATAATGCGGATATACATTTCCAAGTTCAAGCGGATTACTTCATATTTCAACCATACATAGGTGCAAAATTAACTGGTTTAGTTAACAAAAAGTGTTCCACACATTTAAGTGTCCTAGTACATAGAGTTTTTAATGTGGTCATACCACAGCCAACAGAAGAGCCTGGAGACAAGTGGATCGGATTTAGTGTTAAAGAGGGCCAAGAGGTCACTTTTAGAATTGCTGCCTTAGACCTTTACGGAGCTCTTCCTTACATTAGAGGTGAATTAGATGAGAGGAGTCTAAAACTAGAAAACTATGAAATTGATGACGATATTGAAGTTTCTCAACCATTAAATGTACCTTATGTGATTTTTGATAAAACTAAGCCTAGTGTAAAATAA
Protein
MSKIIKFDLRELKQLANDKNSCLVVKKITQNLALQPWCLGNIKESITNLLDYKVGKFDKELNGVLVSYKNPCILQNVGTIRNDNADIHFQVQADYFIFQPYIGAKLTGLVNKKCSTHLSVLVHRVFNVVIPQPTEEPGDKWIGFSVKEGQEVTFRIAALDLYGALPYIRGELDERSLKLENYEIDDDIEVSQPLNVPYVIFDKTKPSVK
Summary
Subunit
Component of the RNA polymerase I (Pol I) complex consisting of at least 13 subunits (By similarity). Interacts with RRN3/TIF-IA.
Similarity
Belongs to the eukaryotic RPA43 RNA polymerase subunit family.
Keywords
Complete proteome
DNA-directed RNA polymerase
Nucleus
Phosphoprotein
Reference proteome
Transcription
Feature
chain DNA-directed RNA polymerase I subunit RPA43
Uniprot
D3JSV9
A0A2H1VKS5
A0A2A4JKN2
A0A1E1W0P4
A0A212EZS8
A0A194QCQ0
+ More
A0A1E1W333 S4PY05 A0A0L7LFB6 A0A067R3D0 A0A1Y1MRS7 A0A232EL26 A0A087ZYI6 A0A0L7QWV4 A0A2A3EB44 U5ETT7 A0A1B6I1S1 A0A310SHG6 A0A023EXR4 A0A0V0G874 A0A0P4VK45 A0A069DZ41 A0A154NZB3 H3AQL8 E9H7S0 E2A3E9 E9J9M9 A0A0M8ZSW5 A0A1B6DAZ5 G7P0S2 F7A560 A0A2K6DJH1 A0A2K5VT47 H9Z6D5 A0A2K6MTI5 A0A2K6QW86 G7MLQ6 H2PMQ7 A0A096NPL9 A0A2J8VEB4 A0A2K6ABF5 A0A2K5MG82 A0A0D9RM70 F6WGE7 A0A2K5JMW9 G9KW12 A0A2Y9DMM2 A0A2K5D818 A0A2K5QMR4 G5BZJ2 A0A2Y9H1J7 A0A2K6SZF5 U6DI23 A0A2R9AZY1 H2QU84 Q3B726 A0A1A8I2S0 U4UTD8 K7CLQ7 G3RU18 W5PJ88 A0A0J7NBY6 A0A3Q0CVH2 A0A2Y9F6A3 A0A2U3X0R0 N6TDJ6 G3T2G1 A0A341AXK8 A0A2I3HSQ3 A0A026W1Z1 A0A1B0GLB8 A0A1A8PXX6 A0A1L8FVY7 A0A0A9ZD88 A0A1S3AB57 Q6GLJ2 A0A1B0D988 A0A2Y9PJS7 A0A2U3V9L8 A0A340X8V4 M3X5H4 A0A1A8CEU4 A0A1A8D5V7 A0A3P4NGT7 M3YR84 H0X025 A0A1A7ZZN7 A0A383YQD0 F1NJS0 A0A1A8HHH6 A0A3L8DM15 A0A091CR13 F7C1U5 A0A158NMM4 A0A2P4TF30
A0A1E1W333 S4PY05 A0A0L7LFB6 A0A067R3D0 A0A1Y1MRS7 A0A232EL26 A0A087ZYI6 A0A0L7QWV4 A0A2A3EB44 U5ETT7 A0A1B6I1S1 A0A310SHG6 A0A023EXR4 A0A0V0G874 A0A0P4VK45 A0A069DZ41 A0A154NZB3 H3AQL8 E9H7S0 E2A3E9 E9J9M9 A0A0M8ZSW5 A0A1B6DAZ5 G7P0S2 F7A560 A0A2K6DJH1 A0A2K5VT47 H9Z6D5 A0A2K6MTI5 A0A2K6QW86 G7MLQ6 H2PMQ7 A0A096NPL9 A0A2J8VEB4 A0A2K6ABF5 A0A2K5MG82 A0A0D9RM70 F6WGE7 A0A2K5JMW9 G9KW12 A0A2Y9DMM2 A0A2K5D818 A0A2K5QMR4 G5BZJ2 A0A2Y9H1J7 A0A2K6SZF5 U6DI23 A0A2R9AZY1 H2QU84 Q3B726 A0A1A8I2S0 U4UTD8 K7CLQ7 G3RU18 W5PJ88 A0A0J7NBY6 A0A3Q0CVH2 A0A2Y9F6A3 A0A2U3X0R0 N6TDJ6 G3T2G1 A0A341AXK8 A0A2I3HSQ3 A0A026W1Z1 A0A1B0GLB8 A0A1A8PXX6 A0A1L8FVY7 A0A0A9ZD88 A0A1S3AB57 Q6GLJ2 A0A1B0D988 A0A2Y9PJS7 A0A2U3V9L8 A0A340X8V4 M3X5H4 A0A1A8CEU4 A0A1A8D5V7 A0A3P4NGT7 M3YR84 H0X025 A0A1A7ZZN7 A0A383YQD0 F1NJS0 A0A1A8HHH6 A0A3L8DM15 A0A091CR13 F7C1U5 A0A158NMM4 A0A2P4TF30
Pubmed
19121390
22118469
26354079
23622113
26227816
24845553
+ More
28004739 28648823 25474469 27129103 26334808 9215903 21292972 20798317 21282665 22002653 17431167 25319552 25362486 23236062 21993625 22722832 16136131 14702039 12690205 15489334 12438708 12393749 17081983 18220336 18669648 20068231 21406692 23186163 23537049 22398555 20809919 28071753 24508170 27762356 25401762 17975172 15592404 30249741 19892987 21347285
28004739 28648823 25474469 27129103 26334808 9215903 21292972 20798317 21282665 22002653 17431167 25319552 25362486 23236062 21993625 22722832 16136131 14702039 12690205 15489334 12438708 12393749 17081983 18220336 18669648 20068231 21406692 23186163 23537049 22398555 20809919 28071753 24508170 27762356 25401762 17975172 15592404 30249741 19892987 21347285
EMBL
BABH01041058
GU288816
ADB78703.1
ODYU01003098
SOQ41423.1
NWSH01001098
+ More
PCG72617.1 GDQN01010583 JAT80471.1 AGBW02011226 OWR47006.1 KQ459232 KPJ02760.1 GDQN01009705 JAT81349.1 GAIX01004128 JAA88432.1 JTDY01001423 KOB73896.1 KK853024 KDR12339.1 GEZM01023394 JAV88412.1 NNAY01003674 OXU19018.1 KQ414710 KOC63019.1 KZ288297 PBC28910.1 GANO01002634 JAB57237.1 GECU01026837 JAS80869.1 KQ766175 OAD53725.1 GBBI01004662 JAC14050.1 GECL01001801 JAP04323.1 GDKW01003749 JAI52846.1 GBGD01001300 JAC87589.1 KQ434785 KZC05016.1 AFYH01032788 AFYH01032789 AFYH01032790 GL732601 EFX72255.1 GL436428 EFN72013.1 GL769399 EFZ10425.1 KQ435912 KOX68859.1 GEDC01014431 JAS22867.1 CM001278 EHH52284.1 JSUE03028627 JSUE03028628 JSUE03028629 AQIA01046942 JU474697 AFH31501.1 JU333411 JV046457 CM001255 AFE77166.1 AFI36528.1 EHH17460.1 ABGA01248136 ABGA01248137 AHZZ02022916 NDHI03003421 PNJ55852.1 AQIB01018527 JP020493 AES09091.1 JH172608 EHB14703.1 HAAF01010806 CCP82630.1 AJFE02088144 AACZ04068668 GABC01010615 GABF01002231 GABE01002144 JAA00723.1 JAA19914.1 JAA42595.1 AK301329 CH236948 CH471073 BC014574 BC130298 BC130300 BK000492 HAED01005180 SBQ91210.1 KB632355 ERL93385.1 GABD01005836 NBAG03000259 JAA27264.1 PNI57982.1 CABD030050087 AMGL01086297 LBMM01007058 KMQ90080.1 APGK01041853 KB740998 ENN75818.1 ADFV01001490 ADFV01001491 KK107485 EZA50082.1 AJWK01034733 HAEG01009962 SBR85867.1 CM004476 OCT75767.1 GBHO01000392 GBRD01005891 JAG43212.1 JAG59930.1 BC074493 AAH74493.1 AJVK01004276 AANG04003224 HADZ01013637 SBP77578.1 HAEA01001145 SBQ29625.1 CYRY02020758 VCW97156.1 AEYP01012949 AEYP01012950 AAQR03057260 HADY01009373 SBP47858.1 AADN05000033 HAEC01014630 SBQ82847.1 QOIP01000006 RLU21421.1 KN124680 KFO20627.1 ADTU01020592 ADTU01020593 PPHD01000992 POI34975.1
PCG72617.1 GDQN01010583 JAT80471.1 AGBW02011226 OWR47006.1 KQ459232 KPJ02760.1 GDQN01009705 JAT81349.1 GAIX01004128 JAA88432.1 JTDY01001423 KOB73896.1 KK853024 KDR12339.1 GEZM01023394 JAV88412.1 NNAY01003674 OXU19018.1 KQ414710 KOC63019.1 KZ288297 PBC28910.1 GANO01002634 JAB57237.1 GECU01026837 JAS80869.1 KQ766175 OAD53725.1 GBBI01004662 JAC14050.1 GECL01001801 JAP04323.1 GDKW01003749 JAI52846.1 GBGD01001300 JAC87589.1 KQ434785 KZC05016.1 AFYH01032788 AFYH01032789 AFYH01032790 GL732601 EFX72255.1 GL436428 EFN72013.1 GL769399 EFZ10425.1 KQ435912 KOX68859.1 GEDC01014431 JAS22867.1 CM001278 EHH52284.1 JSUE03028627 JSUE03028628 JSUE03028629 AQIA01046942 JU474697 AFH31501.1 JU333411 JV046457 CM001255 AFE77166.1 AFI36528.1 EHH17460.1 ABGA01248136 ABGA01248137 AHZZ02022916 NDHI03003421 PNJ55852.1 AQIB01018527 JP020493 AES09091.1 JH172608 EHB14703.1 HAAF01010806 CCP82630.1 AJFE02088144 AACZ04068668 GABC01010615 GABF01002231 GABE01002144 JAA00723.1 JAA19914.1 JAA42595.1 AK301329 CH236948 CH471073 BC014574 BC130298 BC130300 BK000492 HAED01005180 SBQ91210.1 KB632355 ERL93385.1 GABD01005836 NBAG03000259 JAA27264.1 PNI57982.1 CABD030050087 AMGL01086297 LBMM01007058 KMQ90080.1 APGK01041853 KB740998 ENN75818.1 ADFV01001490 ADFV01001491 KK107485 EZA50082.1 AJWK01034733 HAEG01009962 SBR85867.1 CM004476 OCT75767.1 GBHO01000392 GBRD01005891 JAG43212.1 JAG59930.1 BC074493 AAH74493.1 AJVK01004276 AANG04003224 HADZ01013637 SBP77578.1 HAEA01001145 SBQ29625.1 CYRY02020758 VCW97156.1 AEYP01012949 AEYP01012950 AAQR03057260 HADY01009373 SBP47858.1 AADN05000033 HAEC01014630 SBQ82847.1 QOIP01000006 RLU21421.1 KN124680 KFO20627.1 ADTU01020592 ADTU01020593 PPHD01000992 POI34975.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000037510
UP000027135
+ More
UP000215335 UP000005203 UP000053825 UP000242457 UP000076502 UP000008672 UP000000305 UP000000311 UP000053105 UP000009130 UP000006718 UP000233120 UP000233100 UP000233180 UP000233200 UP000001595 UP000028761 UP000233140 UP000233060 UP000029965 UP000008225 UP000233080 UP000248480 UP000233020 UP000233040 UP000006813 UP000248481 UP000233220 UP000240080 UP000002277 UP000005640 UP000030742 UP000001519 UP000002356 UP000036403 UP000189706 UP000248484 UP000245340 UP000019118 UP000007646 UP000252040 UP000001073 UP000053097 UP000092461 UP000186698 UP000079721 UP000092462 UP000248483 UP000245320 UP000265300 UP000011712 UP000000715 UP000005225 UP000261681 UP000000539 UP000279307 UP000028990 UP000002281 UP000005205
UP000215335 UP000005203 UP000053825 UP000242457 UP000076502 UP000008672 UP000000305 UP000000311 UP000053105 UP000009130 UP000006718 UP000233120 UP000233100 UP000233180 UP000233200 UP000001595 UP000028761 UP000233140 UP000233060 UP000029965 UP000008225 UP000233080 UP000248480 UP000233020 UP000233040 UP000006813 UP000248481 UP000233220 UP000240080 UP000002277 UP000005640 UP000030742 UP000001519 UP000002356 UP000036403 UP000189706 UP000248484 UP000245340 UP000019118 UP000007646 UP000252040 UP000001073 UP000053097 UP000092461 UP000186698 UP000079721 UP000092462 UP000248483 UP000245320 UP000265300 UP000011712 UP000000715 UP000005225 UP000261681 UP000000539 UP000279307 UP000028990 UP000002281 UP000005205
Interpro
Gene 3D
ProteinModelPortal
D3JSV9
A0A2H1VKS5
A0A2A4JKN2
A0A1E1W0P4
A0A212EZS8
A0A194QCQ0
+ More
A0A1E1W333 S4PY05 A0A0L7LFB6 A0A067R3D0 A0A1Y1MRS7 A0A232EL26 A0A087ZYI6 A0A0L7QWV4 A0A2A3EB44 U5ETT7 A0A1B6I1S1 A0A310SHG6 A0A023EXR4 A0A0V0G874 A0A0P4VK45 A0A069DZ41 A0A154NZB3 H3AQL8 E9H7S0 E2A3E9 E9J9M9 A0A0M8ZSW5 A0A1B6DAZ5 G7P0S2 F7A560 A0A2K6DJH1 A0A2K5VT47 H9Z6D5 A0A2K6MTI5 A0A2K6QW86 G7MLQ6 H2PMQ7 A0A096NPL9 A0A2J8VEB4 A0A2K6ABF5 A0A2K5MG82 A0A0D9RM70 F6WGE7 A0A2K5JMW9 G9KW12 A0A2Y9DMM2 A0A2K5D818 A0A2K5QMR4 G5BZJ2 A0A2Y9H1J7 A0A2K6SZF5 U6DI23 A0A2R9AZY1 H2QU84 Q3B726 A0A1A8I2S0 U4UTD8 K7CLQ7 G3RU18 W5PJ88 A0A0J7NBY6 A0A3Q0CVH2 A0A2Y9F6A3 A0A2U3X0R0 N6TDJ6 G3T2G1 A0A341AXK8 A0A2I3HSQ3 A0A026W1Z1 A0A1B0GLB8 A0A1A8PXX6 A0A1L8FVY7 A0A0A9ZD88 A0A1S3AB57 Q6GLJ2 A0A1B0D988 A0A2Y9PJS7 A0A2U3V9L8 A0A340X8V4 M3X5H4 A0A1A8CEU4 A0A1A8D5V7 A0A3P4NGT7 M3YR84 H0X025 A0A1A7ZZN7 A0A383YQD0 F1NJS0 A0A1A8HHH6 A0A3L8DM15 A0A091CR13 F7C1U5 A0A158NMM4 A0A2P4TF30
A0A1E1W333 S4PY05 A0A0L7LFB6 A0A067R3D0 A0A1Y1MRS7 A0A232EL26 A0A087ZYI6 A0A0L7QWV4 A0A2A3EB44 U5ETT7 A0A1B6I1S1 A0A310SHG6 A0A023EXR4 A0A0V0G874 A0A0P4VK45 A0A069DZ41 A0A154NZB3 H3AQL8 E9H7S0 E2A3E9 E9J9M9 A0A0M8ZSW5 A0A1B6DAZ5 G7P0S2 F7A560 A0A2K6DJH1 A0A2K5VT47 H9Z6D5 A0A2K6MTI5 A0A2K6QW86 G7MLQ6 H2PMQ7 A0A096NPL9 A0A2J8VEB4 A0A2K6ABF5 A0A2K5MG82 A0A0D9RM70 F6WGE7 A0A2K5JMW9 G9KW12 A0A2Y9DMM2 A0A2K5D818 A0A2K5QMR4 G5BZJ2 A0A2Y9H1J7 A0A2K6SZF5 U6DI23 A0A2R9AZY1 H2QU84 Q3B726 A0A1A8I2S0 U4UTD8 K7CLQ7 G3RU18 W5PJ88 A0A0J7NBY6 A0A3Q0CVH2 A0A2Y9F6A3 A0A2U3X0R0 N6TDJ6 G3T2G1 A0A341AXK8 A0A2I3HSQ3 A0A026W1Z1 A0A1B0GLB8 A0A1A8PXX6 A0A1L8FVY7 A0A0A9ZD88 A0A1S3AB57 Q6GLJ2 A0A1B0D988 A0A2Y9PJS7 A0A2U3V9L8 A0A340X8V4 M3X5H4 A0A1A8CEU4 A0A1A8D5V7 A0A3P4NGT7 M3YR84 H0X025 A0A1A7ZZN7 A0A383YQD0 F1NJS0 A0A1A8HHH6 A0A3L8DM15 A0A091CR13 F7C1U5 A0A158NMM4 A0A2P4TF30
PDB
2RF4
E-value=0.00190789,
Score=94
Ontologies
KEGG
GO
Topology
Subcellular location
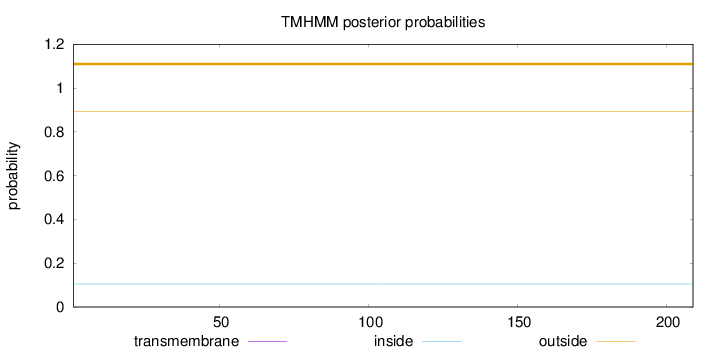
Length:
209
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00494
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.10547
outside
1 - 209
Population Genetic Test Statistics
Pi
134.731947
Theta
154.15315
Tajima's D
-0.75141
CLR
14.741396
CSRT
0.183590820458977
Interpretation
Uncertain
