Gene
KWMTBOMO10319
Pre Gene Modal
BGIBMGA007219
Annotation
PREDICTED:_ubiquitin_carboxyl-terminal_hydrolase_43_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.785
Sequence
CDS
ATGACACAGAAGCAAGTGGAAGGATCCCGATTGAAGCGTACTTTTACTTTACCCAGGAATCCCTTTGGGAACTCAAAGCCTGGTTCAAGTAAAAGTAAAAGTGGCGATGGAGACAACAAATCCATAAATTCTAGTTCTGTGATAAGTGATACACAAAAGGACGAGGGCAGCTTAGATAGGAAGTTGTTCAGAAGGCCTTCATGGAAAAGGTTTCTAAATAAGATAGCTCTACATATGAGTACAGTACGTGTAGCGGGTGGCAAACCGGCTCCAGTTTTAGTTAATGGAGAGAAGACGAATTGTAATGGTGAGCCTCCGTGGCCGCCTGGGAACACTCCAGCTGCGACAGGGATAAAGAACCACGGAAACACTTGTTACATGAATGCTGTCCTGCAGTGCCTTTCTCATACAGATGTTATTGCTGAATATTTTGTGTTGGACCATTATAAGGTTGATCTTCAAAAGCGAAATAAAATTAATTCAAAGAAATATGGAACAAGAGGAGAAGTTACTGAGCAGTTGGCATCATTGTTGAAATCATTATGGGCTTGTCACTACACACCAGATATGAGCACAACCTTTAAGAAAACTGTGGAAAGACATGGCACGCAATATAGAGGTAACAGTCAACATGATGCGCAAGAATTTTTATTTTGGCTTCTTGATAAAGTGCATGAGGATTTAAATACCGCTACAAAAAGAAAATATAAAACAATAAAGAATACTTGTGGGAAGCCAGATGAGGTAGTCGCAGCAGAAACATTAGCAAATCATGCTAGAAGAAATAGCTCATTTGTGCAGACAGTTTTTCAAGCACAATATAGATCGGCCCTAACGTGTGCAAAATGTGAAAGAACATCTTGCACCTTTGACCCCTTCCATTGTGTGAGTGTGCAGCTACCATCAAGATTAGCAACTGCACAACCGTCTCCCCTACCTGTCAATGTAGTTTATGTGAATCAGCAACCTCGACAAGTTCGCATCGGACTGGAGCTACCACCGACCGCAACGATGGATGATTTAAGAACGACACTTCACGCAGATACAGGAATTGAACGCGATCACATTATATTAGCTGAAATCAACGAAGCAGGTTGGTGCAACGCTCGTGCAAACTGGGAAGTTGCTGGTATGGATTTCGGCTCGCTGTATTGTTTAGAGGCTCCGCCATTATTACAAACGCCAACAACTCCATACCTTCTCTTACTATGGGTCAATATAATTGAAGGAGAACGATTTGGTTCACCATATGCAATGCAAGTTCCGCGGGAGATTTCTTATGAAGATTTACAGAAGTTGATCTTGAAGGAAATGAACCAGATTATATCCGAACGAGTTTTGGAAAACGCACAAGGAACTGATATATTCCGCGCGAGGATAGCTGAGGCACATCGACCGCGGGACCCAGCTTATTTGCAACCAGAGCTACCGCATCCATTATTCGCCTTGGACGTAGAGCAAGCCTTGTGTCCGCACGATAAACATCCACATCTCCGGTTGGAACTGCTCTGGGACCCCGAGCAACGTGACAATATAATCCGCGAGTGCAAGGAGCAGTGCGAGGTGCACGTGTCCGCGGGCGGGGGGGGCGCGCCGCCGCTCACGCTGCACGCCTGCCTCACGCACTACACGCGCGCCGAGCGACTGGGACAGGACGACGCCTGGAGATGCCCTCAGTGTCAGAGGTACATGCCTGTGGTAAAAACATTGGGACTGTGGTCACTACCGGACATATTAGTAATACATTTGAAGCGATTTAGACAGCAGCCTAAAGGCCGGACGAGCACGAAATTAACAACAATGGTAGAGTTTCCACTCAACGATTTCGACATGACGCCGCACCTCGCGCACAGGAACTCCTCGGCGGAGTCGCCGGGGCATTCGCGGTCACCACGGCGGAGATACTCAAAAACGTCAACACTGAATCATCACGATAACGTTTACGATTTGTACGCCATTTGTTATCATCACGGCGATGACTTAGAAACAGGTCATTACACTGCCGCCTGCAAAAACCCTTACGACCGCCACTGGTATAAGTTCGACGATTCAAGGGTGACACGGATCGAAGATGAATGTGCCTACGATGAGTTGGTGAACAACACCGCGTACATGCTGTTTTACAAACGGCAGAAGCCGACCGTCACGCACTCCTATAGCTCCGAGGAGTACGGCGGCCATTGGGCGCTTCGCATGCCTAAATACGTAAAGAGGACCACGGAGACATTGAAAGAGCTGACCGAGGTGAAGGAAGAAAACGTCGAAGAAACCAAGATCGATTCGTCGCCCGTCGAACACGACTCTGAGTCTGACGTCACGGCCGTCACGCGCAGCCCTTCGCTCTCCCGGAGCGCGAACAGTCTGCTCGACGACGACGCGCCTCCCGCTAGCAACATCACGACGATAATTCAATCGCCGACCCTACAGAGGCCTCTCATTGTAGAAGTCAATGGAAATAACGTTTGTACAAACGTAAACGAACGCGAGTCTGAGCCGAGCAACTCTATCGAGCTGTATATTCACAAGGACGTGCACGTGAATCCAAAGATGACGCCAGTGGACGGTCGCCGCCCACGCTCCGTGGACATTCCCGTCACGCTCTCTGCACCCCACTCCGCCCCCCTCTCCGCTCCCCACTCCGCCCCCCACTCTGCCCCATTCTGCACTAAAGACTCGAGTAGGAACTATGAGAGCTCCCCCCTAGTCGCGAGCATAAACGAAGTGGAGTACCATCCCACAACTGAAGAGTTGATGATGTCGATGTTTAAAGAATCGCAGTTTATCGTTCCCAGGCGCCCTGATCGGGTCCAAGGAGAATCTCATCATCCAGGAGAAAAGACTTCTGCATGGAAAACTCGAATATCGACATGA
Protein
MTQKQVEGSRLKRTFTLPRNPFGNSKPGSSKSKSGDGDNKSINSSSVISDTQKDEGSLDRKLFRRPSWKRFLNKIALHMSTVRVAGGKPAPVLVNGEKTNCNGEPPWPPGNTPAATGIKNHGNTCYMNAVLQCLSHTDVIAEYFVLDHYKVDLQKRNKINSKKYGTRGEVTEQLASLLKSLWACHYTPDMSTTFKKTVERHGTQYRGNSQHDAQEFLFWLLDKVHEDLNTATKRKYKTIKNTCGKPDEVVAAETLANHARRNSSFVQTVFQAQYRSALTCAKCERTSCTFDPFHCVSVQLPSRLATAQPSPLPVNVVYVNQQPRQVRIGLELPPTATMDDLRTTLHADTGIERDHIILAEINEAGWCNARANWEVAGMDFGSLYCLEAPPLLQTPTTPYLLLLWVNIIEGERFGSPYAMQVPREISYEDLQKLILKEMNQIISERVLENAQGTDIFRARIAEAHRPRDPAYLQPELPHPLFALDVEQALCPHDKHPHLRLELLWDPEQRDNIIRECKEQCEVHVSAGGGGAPPLTLHACLTHYTRAERLGQDDAWRCPQCQRYMPVVKTLGLWSLPDILVIHLKRFRQQPKGRTSTKLTTMVEFPLNDFDMTPHLAHRNSSAESPGHSRSPRRRYSKTSTLNHHDNVYDLYAICYHHGDDLETGHYTAACKNPYDRHWYKFDDSRVTRIEDECAYDELVNNTAYMLFYKRQKPTVTHSYSSEEYGGHWALRMPKYVKRTTETLKELTEVKEENVEETKIDSSPVEHDSESDVTAVTRSPSLSRSANSLLDDDAPPASNITTIIQSPTLQRPLIVEVNGNNVCTNVNERESEPSNSIELYIHKDVHVNPKMTPVDGRRPRSVDIPVTLSAPHSAPLSAPHSAPHSAPFCTKDSSRNYESSPLVASINEVEYHPTTEELMMSMFKESQFIVPRRPDRVQGESHHPGEKTSAWKTRIST
Summary
Similarity
Belongs to the peptidase C19 family.
Uniprot
A0A2A4J9B7
A0A194QCT6
A0A212FK13
A0A194QRM5
A0A1Y1KJY0
A0A1Y1KJX7
+ More
A0A2J7Q3N7 D6WVD6 N6TQG5 A0A0C9R5W2 U4TWS1 A0A1S4EZ18 A0A1Q3F5B0 A0A2A3E429 B0VZX2 A0A087ZQJ2 A0A1B0CQA7 A0A151J1F4 A0A182G2Y3 A0A195AUT5 A0A151JUP2 A0A1L8DTI2 A0A158P108 A0A3L8E4R8 A0A026WEM9 A0A151IP81 A0A0L0CBF2 A0A0L7QPN9 A0A0R3NNL7 A0A182RHT4 A0A182LZ04 A0A0P8Y6C2 A0A084VWY2 A0A0R1DXH3 Q7QJP0 A0A0J9RKD5 A0A310SS87 A0A182J715 A0A1A9Z5L4 Q28Z11 B4GIE9 A0A182XZ64 A0A182JW52 A0A0Q9WAE7 A0A2M4A732 A0A0B4LGJ1 A0A1B0FNL5 A0A1A9XG19 W5JX92 A0A1A9UI57 A0A182UUF3 A0A182P3M8 B3MF42 B4MP85 A0A182WIK6 A0A182UJ30 Q8I1E2 A0A182FI53 A0A151WWA7 A0A0R3NNF6 B4PBT9 A0A182KRQ7 A0A182X3F2 A0A336M563 B4LN45 A0A0J9RKV1 B4QCJ0 A0A182I3L6 Q8I1A0 A0A0M5IX98 B3NQX7 A0A1B6M3H4 A0A0Q9XIQ2 A0A0P8XQX9 Q9W117 A0A1A9WDY2 A0A0Q9W2D8 Q95SG4 B4J4E4 A0A0V0G9K5 A0A1J1I702 A0A023F3Z4 A0A1W4UM30 A0A2H8TYX9 A0A0A9Z165 A0A2S2R6H6 A0A0K8T5F5 A0A0A9YVB5 K7ISH7 J9K6W9 A0A2H8TV79 A0A182NHI6 A0A182QQM7
A0A2J7Q3N7 D6WVD6 N6TQG5 A0A0C9R5W2 U4TWS1 A0A1S4EZ18 A0A1Q3F5B0 A0A2A3E429 B0VZX2 A0A087ZQJ2 A0A1B0CQA7 A0A151J1F4 A0A182G2Y3 A0A195AUT5 A0A151JUP2 A0A1L8DTI2 A0A158P108 A0A3L8E4R8 A0A026WEM9 A0A151IP81 A0A0L0CBF2 A0A0L7QPN9 A0A0R3NNL7 A0A182RHT4 A0A182LZ04 A0A0P8Y6C2 A0A084VWY2 A0A0R1DXH3 Q7QJP0 A0A0J9RKD5 A0A310SS87 A0A182J715 A0A1A9Z5L4 Q28Z11 B4GIE9 A0A182XZ64 A0A182JW52 A0A0Q9WAE7 A0A2M4A732 A0A0B4LGJ1 A0A1B0FNL5 A0A1A9XG19 W5JX92 A0A1A9UI57 A0A182UUF3 A0A182P3M8 B3MF42 B4MP85 A0A182WIK6 A0A182UJ30 Q8I1E2 A0A182FI53 A0A151WWA7 A0A0R3NNF6 B4PBT9 A0A182KRQ7 A0A182X3F2 A0A336M563 B4LN45 A0A0J9RKV1 B4QCJ0 A0A182I3L6 Q8I1A0 A0A0M5IX98 B3NQX7 A0A1B6M3H4 A0A0Q9XIQ2 A0A0P8XQX9 Q9W117 A0A1A9WDY2 A0A0Q9W2D8 Q95SG4 B4J4E4 A0A0V0G9K5 A0A1J1I702 A0A023F3Z4 A0A1W4UM30 A0A2H8TYX9 A0A0A9Z165 A0A2S2R6H6 A0A0K8T5F5 A0A0A9YVB5 K7ISH7 J9K6W9 A0A2H8TV79 A0A182NHI6 A0A182QQM7
Pubmed
26354079
22118469
28004739
18362917
19820115
23537049
+ More
17510324 26483478 21347285 30249741 24508170 26108605 15632085 17994087 24438588 17550304 12364791 22936249 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 20966253 25474469 25401762 20075255
17510324 26483478 21347285 30249741 24508170 26108605 15632085 17994087 24438588 17550304 12364791 22936249 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20920257 23761445 20966253 25474469 25401762 20075255
EMBL
NWSH01002273
PCG68717.1
KQ459232
KPJ02795.1
AGBW02008166
OWR54078.1
+ More
KQ461155 KPJ08142.1 GEZM01081310 JAV61712.1 GEZM01081309 JAV61713.1 NEVH01019065 PNF23197.1 KQ971357 EFA08548.2 APGK01058655 APGK01058656 APGK01058657 KB741291 ENN70541.1 GBYB01008630 GBYB01011678 GBYB01011679 JAG78397.1 JAG81445.1 JAG81446.1 KB631579 ERL84413.1 GFDL01012291 JAV22754.1 KZ288386 PBC26465.1 DS231815 EDS35214.1 AJWK01023339 AJWK01023340 KQ980514 KYN15709.1 JXUM01140331 JXUM01140332 JXUM01140333 KQ569045 KXJ68785.1 KQ976738 KYM75785.1 KQ981732 KYN36579.1 GFDF01004489 JAV09595.1 ADTU01005930 ADTU01005931 QOIP01000001 RLU27453.1 KK107242 EZA54555.1 KQ976878 KYN07649.1 JRES01000655 KNC29550.1 KQ414806 KOC60602.1 CM000071 KRT02567.1 AXCM01000512 CH902619 KPU76965.1 ATLV01017808 KE525192 KFB42476.1 CM000158 KRK00595.1 AAAB01008807 EAA03924.4 CM002911 KMY96418.1 KQ761005 OAD58402.1 EAL25804.3 CH479183 EDW36269.1 CH940648 KRF79207.1 GGFK01003286 MBW36607.1 AE013599 AHN56650.1 AHN56651.1 CCAG010023790 ADMH02000065 ETN67934.1 EDV37670.2 CH963849 EDW73924.1 AY190942 AAO01028.1 KQ982691 KYQ52173.1 KRT02568.1 EDW92593.1 UFQT01000574 SSX25412.1 EDW60049.1 KMY96417.1 CM000362 EDX08606.1 APCN01000215 AY190953 AAO01072.1 CP012524 ALC40894.1 CH954179 EDV57060.1 GEBQ01009491 JAT30486.1 CH933808 KRG04167.1 KPU76966.1 BT057999 AAF47260.2 ACM16709.1 KRF79208.1 AY060809 AAL28357.1 CH916367 EDW01626.1 GECL01001340 JAP04784.1 CVRI01000043 CRK96087.1 GBBI01003033 JAC15679.1 GFXV01007710 MBW19515.1 GBHO01008054 JAG35550.1 GGMS01016433 MBY85636.1 GBRD01005059 JAG60762.1 GBHO01008053 JAG35551.1 ABLF02013269 GFXV01005906 MBW17711.1 AXCN02000594
KQ461155 KPJ08142.1 GEZM01081310 JAV61712.1 GEZM01081309 JAV61713.1 NEVH01019065 PNF23197.1 KQ971357 EFA08548.2 APGK01058655 APGK01058656 APGK01058657 KB741291 ENN70541.1 GBYB01008630 GBYB01011678 GBYB01011679 JAG78397.1 JAG81445.1 JAG81446.1 KB631579 ERL84413.1 GFDL01012291 JAV22754.1 KZ288386 PBC26465.1 DS231815 EDS35214.1 AJWK01023339 AJWK01023340 KQ980514 KYN15709.1 JXUM01140331 JXUM01140332 JXUM01140333 KQ569045 KXJ68785.1 KQ976738 KYM75785.1 KQ981732 KYN36579.1 GFDF01004489 JAV09595.1 ADTU01005930 ADTU01005931 QOIP01000001 RLU27453.1 KK107242 EZA54555.1 KQ976878 KYN07649.1 JRES01000655 KNC29550.1 KQ414806 KOC60602.1 CM000071 KRT02567.1 AXCM01000512 CH902619 KPU76965.1 ATLV01017808 KE525192 KFB42476.1 CM000158 KRK00595.1 AAAB01008807 EAA03924.4 CM002911 KMY96418.1 KQ761005 OAD58402.1 EAL25804.3 CH479183 EDW36269.1 CH940648 KRF79207.1 GGFK01003286 MBW36607.1 AE013599 AHN56650.1 AHN56651.1 CCAG010023790 ADMH02000065 ETN67934.1 EDV37670.2 CH963849 EDW73924.1 AY190942 AAO01028.1 KQ982691 KYQ52173.1 KRT02568.1 EDW92593.1 UFQT01000574 SSX25412.1 EDW60049.1 KMY96417.1 CM000362 EDX08606.1 APCN01000215 AY190953 AAO01072.1 CP012524 ALC40894.1 CH954179 EDV57060.1 GEBQ01009491 JAT30486.1 CH933808 KRG04167.1 KPU76966.1 BT057999 AAF47260.2 ACM16709.1 KRF79208.1 AY060809 AAL28357.1 CH916367 EDW01626.1 GECL01001340 JAP04784.1 CVRI01000043 CRK96087.1 GBBI01003033 JAC15679.1 GFXV01007710 MBW19515.1 GBHO01008054 JAG35550.1 GGMS01016433 MBY85636.1 GBRD01005059 JAG60762.1 GBHO01008053 JAG35551.1 ABLF02013269 GFXV01005906 MBW17711.1 AXCN02000594
Proteomes
UP000218220
UP000053268
UP000007151
UP000053240
UP000235965
UP000007266
+ More
UP000019118 UP000030742 UP000242457 UP000002320 UP000005203 UP000092461 UP000078492 UP000069940 UP000249989 UP000078540 UP000078541 UP000005205 UP000279307 UP000053097 UP000078542 UP000037069 UP000053825 UP000001819 UP000075900 UP000075883 UP000007801 UP000030765 UP000002282 UP000007062 UP000075880 UP000092445 UP000008744 UP000076408 UP000075881 UP000008792 UP000000803 UP000092444 UP000092443 UP000000673 UP000078200 UP000075903 UP000075885 UP000007798 UP000075920 UP000075902 UP000069272 UP000075809 UP000075882 UP000076407 UP000000304 UP000075840 UP000092553 UP000008711 UP000009192 UP000091820 UP000001070 UP000183832 UP000192221 UP000002358 UP000007819 UP000075884 UP000075886
UP000019118 UP000030742 UP000242457 UP000002320 UP000005203 UP000092461 UP000078492 UP000069940 UP000249989 UP000078540 UP000078541 UP000005205 UP000279307 UP000053097 UP000078542 UP000037069 UP000053825 UP000001819 UP000075900 UP000075883 UP000007801 UP000030765 UP000002282 UP000007062 UP000075880 UP000092445 UP000008744 UP000076408 UP000075881 UP000008792 UP000000803 UP000092444 UP000092443 UP000000673 UP000078200 UP000075903 UP000075885 UP000007798 UP000075920 UP000075902 UP000069272 UP000075809 UP000075882 UP000076407 UP000000304 UP000075840 UP000092553 UP000008711 UP000009192 UP000091820 UP000001070 UP000183832 UP000192221 UP000002358 UP000007819 UP000075884 UP000075886
Interpro
SUPFAM
SSF54001
SSF54001
ProteinModelPortal
A0A2A4J9B7
A0A194QCT6
A0A212FK13
A0A194QRM5
A0A1Y1KJY0
A0A1Y1KJX7
+ More
A0A2J7Q3N7 D6WVD6 N6TQG5 A0A0C9R5W2 U4TWS1 A0A1S4EZ18 A0A1Q3F5B0 A0A2A3E429 B0VZX2 A0A087ZQJ2 A0A1B0CQA7 A0A151J1F4 A0A182G2Y3 A0A195AUT5 A0A151JUP2 A0A1L8DTI2 A0A158P108 A0A3L8E4R8 A0A026WEM9 A0A151IP81 A0A0L0CBF2 A0A0L7QPN9 A0A0R3NNL7 A0A182RHT4 A0A182LZ04 A0A0P8Y6C2 A0A084VWY2 A0A0R1DXH3 Q7QJP0 A0A0J9RKD5 A0A310SS87 A0A182J715 A0A1A9Z5L4 Q28Z11 B4GIE9 A0A182XZ64 A0A182JW52 A0A0Q9WAE7 A0A2M4A732 A0A0B4LGJ1 A0A1B0FNL5 A0A1A9XG19 W5JX92 A0A1A9UI57 A0A182UUF3 A0A182P3M8 B3MF42 B4MP85 A0A182WIK6 A0A182UJ30 Q8I1E2 A0A182FI53 A0A151WWA7 A0A0R3NNF6 B4PBT9 A0A182KRQ7 A0A182X3F2 A0A336M563 B4LN45 A0A0J9RKV1 B4QCJ0 A0A182I3L6 Q8I1A0 A0A0M5IX98 B3NQX7 A0A1B6M3H4 A0A0Q9XIQ2 A0A0P8XQX9 Q9W117 A0A1A9WDY2 A0A0Q9W2D8 Q95SG4 B4J4E4 A0A0V0G9K5 A0A1J1I702 A0A023F3Z4 A0A1W4UM30 A0A2H8TYX9 A0A0A9Z165 A0A2S2R6H6 A0A0K8T5F5 A0A0A9YVB5 K7ISH7 J9K6W9 A0A2H8TV79 A0A182NHI6 A0A182QQM7
A0A2J7Q3N7 D6WVD6 N6TQG5 A0A0C9R5W2 U4TWS1 A0A1S4EZ18 A0A1Q3F5B0 A0A2A3E429 B0VZX2 A0A087ZQJ2 A0A1B0CQA7 A0A151J1F4 A0A182G2Y3 A0A195AUT5 A0A151JUP2 A0A1L8DTI2 A0A158P108 A0A3L8E4R8 A0A026WEM9 A0A151IP81 A0A0L0CBF2 A0A0L7QPN9 A0A0R3NNL7 A0A182RHT4 A0A182LZ04 A0A0P8Y6C2 A0A084VWY2 A0A0R1DXH3 Q7QJP0 A0A0J9RKD5 A0A310SS87 A0A182J715 A0A1A9Z5L4 Q28Z11 B4GIE9 A0A182XZ64 A0A182JW52 A0A0Q9WAE7 A0A2M4A732 A0A0B4LGJ1 A0A1B0FNL5 A0A1A9XG19 W5JX92 A0A1A9UI57 A0A182UUF3 A0A182P3M8 B3MF42 B4MP85 A0A182WIK6 A0A182UJ30 Q8I1E2 A0A182FI53 A0A151WWA7 A0A0R3NNF6 B4PBT9 A0A182KRQ7 A0A182X3F2 A0A336M563 B4LN45 A0A0J9RKV1 B4QCJ0 A0A182I3L6 Q8I1A0 A0A0M5IX98 B3NQX7 A0A1B6M3H4 A0A0Q9XIQ2 A0A0P8XQX9 Q9W117 A0A1A9WDY2 A0A0Q9W2D8 Q95SG4 B4J4E4 A0A0V0G9K5 A0A1J1I702 A0A023F3Z4 A0A1W4UM30 A0A2H8TYX9 A0A0A9Z165 A0A2S2R6H6 A0A0K8T5F5 A0A0A9YVB5 K7ISH7 J9K6W9 A0A2H8TV79 A0A182NHI6 A0A182QQM7
PDB
2Y6E
E-value=5.35736e-38,
Score=399
Ontologies
GO
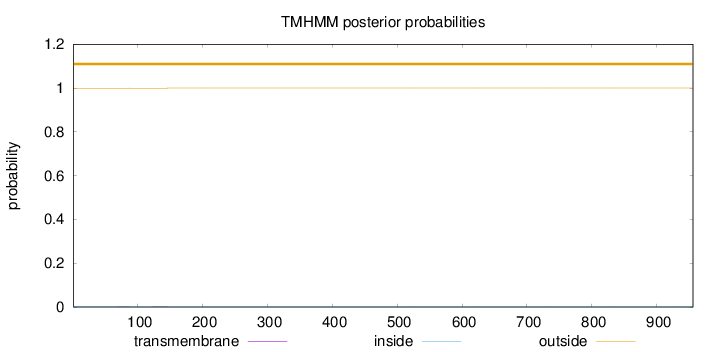
Topology
Length:
956
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01527
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00075
outside
1 - 956
Population Genetic Test Statistics
Pi
165.62966
Theta
150.845322
Tajima's D
0.490583
CLR
0.782804
CSRT
0.507224638768062
Interpretation
Uncertain
