Gene
KWMTBOMO10309
Pre Gene Modal
BGIBMGA007259
Annotation
L-xylulose_reductase_[Bombyx_mori]
Full name
L-xylulose reductase
Alternative Name
Dicarbonyl/L-xylulose reductase
Sperm antigen P26h
Sperm antigen P26h
Location in the cell
Cytoplasmic Reliability : 1.929
Sequence
CDS
ATGGATAAATATTTTGAAGGAAAAAGAATACTTGTTACTGGTGGTTGTCAAGGGATAGGTCGTGGATTAGTCTTAGAACTTTGGAGACTGGGTGCGAATGTTGTTACTGTTTCAAATAATGCTGAAAACTTAAACAAACTCAAAGAGGAATATCCATCCATTGAAATAGTGTACGTTGATCTTCGAAATTGGAATAAAACAAGAGAAGTCATTGATGCTCTTGGAGTTTTTGATGGCCTTGTCAATAATGCCGGTATTGCTATAATCGAACCTTTTTTGGAATGCAAAAAAGAAAGCCTAAATGAAACCTTTGATGTAAATGTTAATGCCGTTATTAATGTGAGTCAAGTGGTAGCGAAAAAAATGATAGAAAACAAAAGAAAAGGTTCCATCGTAAATATTTCATCGCAAGCTTCTAAGGCAGCTCTAAAGGATCACGTAGCATATTGTGCCTCAAAAGGTGCTCTAGACGCACTAACCAGGGTTATGGCCCTAGAACTGGGACCTTTTGGCATACGAATTAATACAGTTAACCCTACTGTCATTATGACTGAAATGGGTAGAAAAGTTTGGGCGGAACCATCAAAATCAAAGGAAATGTTATCTAAAATACCTCTTGGAAGATTCGGTGAGGTGGAGGAAGTGGTTAATGCGGTAATATTCCTGCTTTGTGACAGTGCTAGTTTAATTTCTGGAGTACAATTACCAATTGACGGAGGGTTTTTGGCAACATAA
Protein
MDKYFEGKRILVTGGCQGIGRGLVLELWRLGANVVTVSNNAENLNKLKEEYPSIEIVYVDLRNWNKTREVIDALGVFDGLVNNAGIAIIEPFLECKKESLNETFDVNVNAVINVSQVVAKKMIENKRKGSIVNISSQASKAALKDHVAYCASKGALDALTRVMALELGPFGIRINTVNPTVIMTEMGRKVWAEPSKSKEMLSKIPLGRFGEVEEVVNAVIFLLCDSASLISGVQLPIDGGFLAT
Summary
Catalytic Activity
NADP(+) + xylitol = H(+) + L-xylulose + NADPH
Biophysicochemical Properties
0.11 mM for L-xylulose (at 25 degrees Celsius)
Subunit
Homotetramer.
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Keywords
Acetylation
Carbohydrate metabolism
Complete proteome
Cytoplasmic vesicle
Direct protein sequencing
Glucose metabolism
Membrane
Methylation
NADP
Oxidoreductase
Phosphoprotein
Reference proteome
Xylose metabolism
Cell membrane
Feature
chain L-xylulose reductase
Uniprot
H9JCL4
Q1HPW6
A0A194QBJ4
A0A2H1WQM2
A0A2A4JDX1
A0A194QRL3
+ More
A0A2H1WQD7 A0A212EMP4 A0A194QCS6 A0A194QT21 A0A212EMJ1 S4NJ55 A0A3S2NR58 A0A194QRN6 A0A2A4JCF4 A0A194QBF1 H9JCL3 A0A0L7LCK1 I4DPS3 A0A212ET48 A0A0B4J2T8 A0A1B6KAD0 A0A1B6KQK9 A0A3L8DFD7 A0A0T6BAL7 A0A1B6CL77 A0A1B6HKU2 A0A0B4J2P7 A0A1B6EM10 A0A0L7QQ52 H3AXF4 A0A3S2MIQ7 A0A067QQF0 A0A1B6E6K6 A0A3P9I215 A0A2J7PLW4 G1KJT4 E9FWF2 A0A3P9L555 A0A3B3HPT7 A0A232F2W7 A0A3B4DUY0 A0A1W4WDN7 A0A3Q2ZWE8 C3KGY6 A0A1A7XQ15 A0A1A7YJ59 Q567K5 A0A060Y4P2 A0A3Q1BGC9 G3GXD0 B5X5V8 A0A1D2MSF7 A0A3Q1JRU4 A0A3P8YKW3 A0A3Q1H4K4 A0A0A9XW67 A0A3B4YYI2 A0A3Q3LTD3 Q6P630 A0A3B3DJP1 F6ZAZ2 A0A1U7Q257 Q91XV4 A0A3Q0R148 A0A287D7E0 A0A3Q3N2A4 A0A3Q4N9X8 A0A287D9C5 A0A3B4XW07 A0A3Q2VTJ7 Q4QR61 A0A3B4GQP7 A0A3B4UVP5 A0A1A8M8Y6 A0A1A8H172 A0A1A8DRS3 A0A1U7SVA7 F4WJ19 A0A286ZQ44 E2B5S7 A0A3P8T7Q8 A0A0B7B8J0 A0A1A8A5C5 A0A1A8U1F4 Q920P0 A0A1A8K3X2 A0A1W5AWA2 Q91X52 A0A1S2ZU09 A0A026WNB0 A0A1A8Q5H3 E2ARD3 A0A1A8FCA1 A0A3Q3J7V8 A0A3P8N8H7 A0A3P9DCU8 A0A3B1IVZ8 M3WFI7 A0A2Y9PQ94
A0A2H1WQD7 A0A212EMP4 A0A194QCS6 A0A194QT21 A0A212EMJ1 S4NJ55 A0A3S2NR58 A0A194QRN6 A0A2A4JCF4 A0A194QBF1 H9JCL3 A0A0L7LCK1 I4DPS3 A0A212ET48 A0A0B4J2T8 A0A1B6KAD0 A0A1B6KQK9 A0A3L8DFD7 A0A0T6BAL7 A0A1B6CL77 A0A1B6HKU2 A0A0B4J2P7 A0A1B6EM10 A0A0L7QQ52 H3AXF4 A0A3S2MIQ7 A0A067QQF0 A0A1B6E6K6 A0A3P9I215 A0A2J7PLW4 G1KJT4 E9FWF2 A0A3P9L555 A0A3B3HPT7 A0A232F2W7 A0A3B4DUY0 A0A1W4WDN7 A0A3Q2ZWE8 C3KGY6 A0A1A7XQ15 A0A1A7YJ59 Q567K5 A0A060Y4P2 A0A3Q1BGC9 G3GXD0 B5X5V8 A0A1D2MSF7 A0A3Q1JRU4 A0A3P8YKW3 A0A3Q1H4K4 A0A0A9XW67 A0A3B4YYI2 A0A3Q3LTD3 Q6P630 A0A3B3DJP1 F6ZAZ2 A0A1U7Q257 Q91XV4 A0A3Q0R148 A0A287D7E0 A0A3Q3N2A4 A0A3Q4N9X8 A0A287D9C5 A0A3B4XW07 A0A3Q2VTJ7 Q4QR61 A0A3B4GQP7 A0A3B4UVP5 A0A1A8M8Y6 A0A1A8H172 A0A1A8DRS3 A0A1U7SVA7 F4WJ19 A0A286ZQ44 E2B5S7 A0A3P8T7Q8 A0A0B7B8J0 A0A1A8A5C5 A0A1A8U1F4 Q920P0 A0A1A8K3X2 A0A1W5AWA2 Q91X52 A0A1S2ZU09 A0A026WNB0 A0A1A8Q5H3 E2ARD3 A0A1A8FCA1 A0A3Q3J7V8 A0A3P8N8H7 A0A3P9DCU8 A0A3B1IVZ8 M3WFI7 A0A2Y9PQ94
EC Number
1.1.1.10
Pubmed
19121390
26354079
22118469
23622113
26227816
22651552
+ More
20075255 30249741 9215903 24845553 17554307 21292972 28648823 23594743 24755649 21804562 23929341 29704459 20433749 27289101 25069045 25401762 26823975 29451363 19892987 9890754 11306103 11882650 25186727 27762356 21719571 30723633 20798317 12604240 21477590 16141072 15489334 21183079 24508170 25329095 17975172
20075255 30249741 9215903 24845553 17554307 21292972 28648823 23594743 24755649 21804562 23929341 29704459 20433749 27289101 25069045 25401762 26823975 29451363 19892987 9890754 11306103 11882650 25186727 27762356 21719571 30723633 20798317 12604240 21477590 16141072 15489334 21183079 24508170 25329095 17975172
EMBL
BABH01034711
DQ443286
ABF51375.1
KQ459232
KPJ02784.1
ODYU01010271
+ More
SOQ55262.1 NWSH01001898 PCG69754.1 KQ461155 KPJ08153.1 SOQ55263.1 AGBW02013813 OWR42711.1 KPJ02785.1 KPJ08150.1 OWR42710.1 GAIX01013814 JAA78746.1 RSAL01000006 RVE54201.1 KPJ08152.1 PCG69755.1 KPJ02787.1 BABH01034712 JTDY01001666 KOB73208.1 AK403751 BAM19913.1 AGBW02012635 OWR42712.1 OWR44673.1 GEBQ01031577 GEBQ01023032 JAT08400.1 JAT16945.1 GEBQ01026252 JAT13725.1 QOIP01000009 RLU18873.1 LJIG01003013 KRT83927.1 GEDC01023843 GEDC01023079 GEDC01020713 JAS13455.1 JAS14219.1 JAS16585.1 GECU01036917 GECU01032419 GECU01026666 GECU01001796 JAS70789.1 JAS75287.1 JAS81040.1 JAT05911.1 GECZ01030802 JAS38967.1 KQ414806 KOC60616.1 AFYH01016877 AFYH01016878 AFYH01016879 AFYH01016880 CM012455 RVE60060.1 KK853134 KDR10850.1 GEDC01003718 JAS33580.1 NEVH01024423 PNF17309.1 GL732526 EFX87881.1 NNAY01001163 OXU24932.1 BT082201 ACQ57908.1 HADW01018500 SBP19900.1 HADX01008006 SBP30238.1 BX548040 BC093141 AAH93141.1 FR907432 CDQ86686.1 JH000058 KE682086 RAZU01000244 EGW06279.1 ERE67862.1 RLQ62673.1 BT046427 BT058382 BT059117 BT060083 BT125410 ACI66228.1 ACN10095.1 ACN10830.1 ADM16151.1 LJIJ01000639 ODM95655.1 GBHO01019978 GBHO01019977 GBHO01019976 GBRD01015772 GDHC01019453 JAG23626.1 JAG23627.1 JAG23628.1 JAG50054.1 JAP99175.1 BC062504 CR762052 AAH62504.1 CAJ81504.1 AB045204 AGTP01070806 BC097532 CM004482 AAH97532.1 OCT62589.1 HAEF01011660 HAEG01001859 SBR52769.1 HAEC01009022 SBQ77238.1 HAEA01007778 SBQ36258.1 GL888181 EGI65789.1 AEMK02000080 DQIR01193138 HDB48615.1 GL445887 EFN88896.1 HACG01042778 HACG01042779 CEK89643.1 CEK89644.1 HADY01011393 SBP49878.1 HAEJ01001448 SBS41905.1 AB061719 HAEE01007070 SBR27090.1 D89656 AK004023 AK007627 AK153521 BC012247 KK107148 EZA57458.1 HAEH01010174 SBR89050.1 GL442047 EFN64011.1 HAEB01009909 SBQ56436.1 AANG04004184
SOQ55262.1 NWSH01001898 PCG69754.1 KQ461155 KPJ08153.1 SOQ55263.1 AGBW02013813 OWR42711.1 KPJ02785.1 KPJ08150.1 OWR42710.1 GAIX01013814 JAA78746.1 RSAL01000006 RVE54201.1 KPJ08152.1 PCG69755.1 KPJ02787.1 BABH01034712 JTDY01001666 KOB73208.1 AK403751 BAM19913.1 AGBW02012635 OWR42712.1 OWR44673.1 GEBQ01031577 GEBQ01023032 JAT08400.1 JAT16945.1 GEBQ01026252 JAT13725.1 QOIP01000009 RLU18873.1 LJIG01003013 KRT83927.1 GEDC01023843 GEDC01023079 GEDC01020713 JAS13455.1 JAS14219.1 JAS16585.1 GECU01036917 GECU01032419 GECU01026666 GECU01001796 JAS70789.1 JAS75287.1 JAS81040.1 JAT05911.1 GECZ01030802 JAS38967.1 KQ414806 KOC60616.1 AFYH01016877 AFYH01016878 AFYH01016879 AFYH01016880 CM012455 RVE60060.1 KK853134 KDR10850.1 GEDC01003718 JAS33580.1 NEVH01024423 PNF17309.1 GL732526 EFX87881.1 NNAY01001163 OXU24932.1 BT082201 ACQ57908.1 HADW01018500 SBP19900.1 HADX01008006 SBP30238.1 BX548040 BC093141 AAH93141.1 FR907432 CDQ86686.1 JH000058 KE682086 RAZU01000244 EGW06279.1 ERE67862.1 RLQ62673.1 BT046427 BT058382 BT059117 BT060083 BT125410 ACI66228.1 ACN10095.1 ACN10830.1 ADM16151.1 LJIJ01000639 ODM95655.1 GBHO01019978 GBHO01019977 GBHO01019976 GBRD01015772 GDHC01019453 JAG23626.1 JAG23627.1 JAG23628.1 JAG50054.1 JAP99175.1 BC062504 CR762052 AAH62504.1 CAJ81504.1 AB045204 AGTP01070806 BC097532 CM004482 AAH97532.1 OCT62589.1 HAEF01011660 HAEG01001859 SBR52769.1 HAEC01009022 SBQ77238.1 HAEA01007778 SBQ36258.1 GL888181 EGI65789.1 AEMK02000080 DQIR01193138 HDB48615.1 GL445887 EFN88896.1 HACG01042778 HACG01042779 CEK89643.1 CEK89644.1 HADY01011393 SBP49878.1 HAEJ01001448 SBS41905.1 AB061719 HAEE01007070 SBR27090.1 D89656 AK004023 AK007627 AK153521 BC012247 KK107148 EZA57458.1 HAEH01010174 SBR89050.1 GL442047 EFN64011.1 HAEB01009909 SBQ56436.1 AANG04004184
Proteomes
UP000005204
UP000053268
UP000218220
UP000053240
UP000007151
UP000283053
+ More
UP000037510 UP000002358 UP000279307 UP000005203 UP000053825 UP000008672 UP000027135 UP000265200 UP000235965 UP000001646 UP000000305 UP000265180 UP000001038 UP000215335 UP000261440 UP000192223 UP000264800 UP000000437 UP000193380 UP000257160 UP000001075 UP000030759 UP000273346 UP000087266 UP000094527 UP000265040 UP000265140 UP000257200 UP000261400 UP000261660 UP000261560 UP000002281 UP000189706 UP000261340 UP000005215 UP000261640 UP000261580 UP000261360 UP000264840 UP000186698 UP000261460 UP000261420 UP000189704 UP000007755 UP000008227 UP000008237 UP000265080 UP000002494 UP000192224 UP000000589 UP000079721 UP000053097 UP000000311 UP000261600 UP000265100 UP000265160 UP000018467 UP000011712 UP000248483
UP000037510 UP000002358 UP000279307 UP000005203 UP000053825 UP000008672 UP000027135 UP000265200 UP000235965 UP000001646 UP000000305 UP000265180 UP000001038 UP000215335 UP000261440 UP000192223 UP000264800 UP000000437 UP000193380 UP000257160 UP000001075 UP000030759 UP000273346 UP000087266 UP000094527 UP000265040 UP000265140 UP000257200 UP000261400 UP000261660 UP000261560 UP000002281 UP000189706 UP000261340 UP000005215 UP000261640 UP000261580 UP000261360 UP000264840 UP000186698 UP000261460 UP000261420 UP000189704 UP000007755 UP000008227 UP000008237 UP000265080 UP000002494 UP000192224 UP000000589 UP000079721 UP000053097 UP000000311 UP000261600 UP000265100 UP000265160 UP000018467 UP000011712 UP000248483
Interpro
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9JCL4
Q1HPW6
A0A194QBJ4
A0A2H1WQM2
A0A2A4JDX1
A0A194QRL3
+ More
A0A2H1WQD7 A0A212EMP4 A0A194QCS6 A0A194QT21 A0A212EMJ1 S4NJ55 A0A3S2NR58 A0A194QRN6 A0A2A4JCF4 A0A194QBF1 H9JCL3 A0A0L7LCK1 I4DPS3 A0A212ET48 A0A0B4J2T8 A0A1B6KAD0 A0A1B6KQK9 A0A3L8DFD7 A0A0T6BAL7 A0A1B6CL77 A0A1B6HKU2 A0A0B4J2P7 A0A1B6EM10 A0A0L7QQ52 H3AXF4 A0A3S2MIQ7 A0A067QQF0 A0A1B6E6K6 A0A3P9I215 A0A2J7PLW4 G1KJT4 E9FWF2 A0A3P9L555 A0A3B3HPT7 A0A232F2W7 A0A3B4DUY0 A0A1W4WDN7 A0A3Q2ZWE8 C3KGY6 A0A1A7XQ15 A0A1A7YJ59 Q567K5 A0A060Y4P2 A0A3Q1BGC9 G3GXD0 B5X5V8 A0A1D2MSF7 A0A3Q1JRU4 A0A3P8YKW3 A0A3Q1H4K4 A0A0A9XW67 A0A3B4YYI2 A0A3Q3LTD3 Q6P630 A0A3B3DJP1 F6ZAZ2 A0A1U7Q257 Q91XV4 A0A3Q0R148 A0A287D7E0 A0A3Q3N2A4 A0A3Q4N9X8 A0A287D9C5 A0A3B4XW07 A0A3Q2VTJ7 Q4QR61 A0A3B4GQP7 A0A3B4UVP5 A0A1A8M8Y6 A0A1A8H172 A0A1A8DRS3 A0A1U7SVA7 F4WJ19 A0A286ZQ44 E2B5S7 A0A3P8T7Q8 A0A0B7B8J0 A0A1A8A5C5 A0A1A8U1F4 Q920P0 A0A1A8K3X2 A0A1W5AWA2 Q91X52 A0A1S2ZU09 A0A026WNB0 A0A1A8Q5H3 E2ARD3 A0A1A8FCA1 A0A3Q3J7V8 A0A3P8N8H7 A0A3P9DCU8 A0A3B1IVZ8 M3WFI7 A0A2Y9PQ94
A0A2H1WQD7 A0A212EMP4 A0A194QCS6 A0A194QT21 A0A212EMJ1 S4NJ55 A0A3S2NR58 A0A194QRN6 A0A2A4JCF4 A0A194QBF1 H9JCL3 A0A0L7LCK1 I4DPS3 A0A212ET48 A0A0B4J2T8 A0A1B6KAD0 A0A1B6KQK9 A0A3L8DFD7 A0A0T6BAL7 A0A1B6CL77 A0A1B6HKU2 A0A0B4J2P7 A0A1B6EM10 A0A0L7QQ52 H3AXF4 A0A3S2MIQ7 A0A067QQF0 A0A1B6E6K6 A0A3P9I215 A0A2J7PLW4 G1KJT4 E9FWF2 A0A3P9L555 A0A3B3HPT7 A0A232F2W7 A0A3B4DUY0 A0A1W4WDN7 A0A3Q2ZWE8 C3KGY6 A0A1A7XQ15 A0A1A7YJ59 Q567K5 A0A060Y4P2 A0A3Q1BGC9 G3GXD0 B5X5V8 A0A1D2MSF7 A0A3Q1JRU4 A0A3P8YKW3 A0A3Q1H4K4 A0A0A9XW67 A0A3B4YYI2 A0A3Q3LTD3 Q6P630 A0A3B3DJP1 F6ZAZ2 A0A1U7Q257 Q91XV4 A0A3Q0R148 A0A287D7E0 A0A3Q3N2A4 A0A3Q4N9X8 A0A287D9C5 A0A3B4XW07 A0A3Q2VTJ7 Q4QR61 A0A3B4GQP7 A0A3B4UVP5 A0A1A8M8Y6 A0A1A8H172 A0A1A8DRS3 A0A1U7SVA7 F4WJ19 A0A286ZQ44 E2B5S7 A0A3P8T7Q8 A0A0B7B8J0 A0A1A8A5C5 A0A1A8U1F4 Q920P0 A0A1A8K3X2 A0A1W5AWA2 Q91X52 A0A1S2ZU09 A0A026WNB0 A0A1A8Q5H3 E2ARD3 A0A1A8FCA1 A0A3Q3J7V8 A0A3P8N8H7 A0A3P9DCU8 A0A3B1IVZ8 M3WFI7 A0A2Y9PQ94
PDB
3D3W
E-value=1.51815e-66,
Score=639
Ontologies
PATHWAY
GO
GO:0016491
GO:0050662
GO:0051289
GO:0005881
GO:0042802
GO:0050038
GO:0005997
GO:0006006
GO:0016655
GO:0004090
GO:0005903
GO:0006739
GO:0005902
GO:0044105
GO:0042732
GO:0001669
GO:0016020
GO:0055114
GO:0016324
GO:0016614
GO:0005524
GO:0006468
GO:0016021
GO:0015935
GO:0004812
GO:0006418
GO:0003723
GO:0046872
GO:0003824
Topology
Subcellular location
Membrane
Cytoplasmic vesicle
Secretory vesicle
Acrosome
Apical cell membrane Probably recruited to membranes via an interaction with phosphatidylinositol (By similarity). In kidney, it is localized in the brush border membranes of proximal tubular cells. With evidence from 1 publications.
Cytoplasmic vesicle
Secretory vesicle
Acrosome
Apical cell membrane Probably recruited to membranes via an interaction with phosphatidylinositol (By similarity). In kidney, it is localized in the brush border membranes of proximal tubular cells. With evidence from 1 publications.
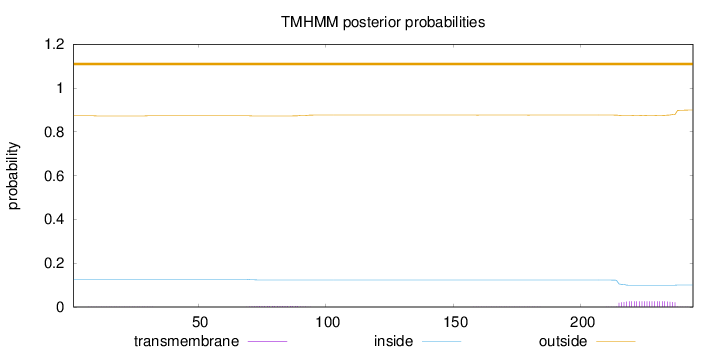
Length:
244
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.6816
Exp number, first 60 AAs:
0.01839
Total prob of N-in:
0.12672
outside
1 - 244
Population Genetic Test Statistics
Pi
176.510975
Theta
175.108587
Tajima's D
0.212419
CLR
0
CSRT
0.427628618569072
Interpretation
Uncertain
