Gene
KWMTBOMO10304
Pre Gene Modal
BGIBMGA007301
Annotation
PREDICTED:_nudix_hydrolase_8-like_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.164
Sequence
CDS
ATGCTTTTCATAGCCACTGAAGACAGATATAATGGCCTTACAATTGAGGACACAGAAAAATCAACAGAAATTGAATTTAACGCTAAGCTAGACTATTCCATAAATAAATGGAAATCAGAAGGCCGAAAATGTTTATGGTTTAAAATAAATATAAAAAATTCCGTTTATGTTCCATTGTTAGCACAAAAAGGTTTTAATTTTCATCATGCCAGAGATGACTTTGTGATGATGTACAAATGGTTACCTGATGACTCCGAACCAAATCTACCTCCTGCCTGTCACACAAATTTGGGTGTTGGTGCCTTAGTCTTTAACAGAAAAAACCAGATGTTGGCTATTTCAGAAAAACATTACGAATACCCACATTGGAAATTACCTGGAGGATATGTTGAGAAAGGGGAAGATATTGTAGACGCAGCGATCAGAGAAGTCAAAGAAGAAACTGGTGTAGACGTGTCATTTTTGTCTTTGATTACACTTCGACACTCGCATAATATGATGTACGGAAACTCGGACATTTACTTACTGCTCATGATGAACGCAATCTCAGATGAGATTACTCTATCTCAGAGAGAAGTCAAAGATTGTAAGTGGATGGATATTAAAGAATACACCACTCATCCTCATGTACACGAATTTAACAGGTTGATTGTGAGAAAAGCCCTCGATTACAAAACAAGAAATATAAAACTCGACATACAGAAGCGTACAGTGAAATGGTCACAGTTTGTTAGAGAAATGAATATATTATCCCTCGAGGACTATAATTACCATTGA
Protein
MLFIATEDRYNGLTIEDTEKSTEIEFNAKLDYSINKWKSEGRKCLWFKINIKNSVYVPLLAQKGFNFHHARDDFVMMYKWLPDDSEPNLPPACHTNLGVGALVFNRKNQMLAISEKHYEYPHWKLPGGYVEKGEDIVDAAIREVKEETGVDVSFLSLITLRHSHNMMYGNSDIYLLLMMNAISDEITLSQREVKDCKWMDIKEYTTHPHVHEFNRLIVRKALDYKTRNIKLDIQKRTVKWSQFVREMNILSLEDYNYH
Summary
Similarity
Belongs to the Nudix hydrolase family.
Uniprot
A0A2A4JMD5
H9JCQ4
A0A3S2P0X8
A0A2H1WQU9
A0A194QH61
A0A194QT27
+ More
A0A212EMJ0 A0A2H1WQU5 H9JCL6 A0A194QCS1 A0A2A4JMV9 A0A212FA52 A0A194QX54 D6WX88 A0A1W4WIP4 A0A1W4WUU4 A0A1W4WTJ0 A0A0T6AX11 S4P8F4 A0A2J7PKN1 A0A067R6N0 N6U4S0 U4UAU5 A0A2J7PKL9 A0A182RB96 A0A1J1I3V7 A0A182P406 A0A023ENS3 A0A182M9X4 A0A2M3Z6F6 A0A182SS98 A0A2M3Z695 A0A2M3Z627 A0A2M3Z5Y8 A0A3R7MJ34 A0A182H9K5 A0A182YG36 A0A182W106 A0A182ND52 A0A336K395 T1DR80 U5EQK4 A0A2M4BTW3 A0A2M4BTC5 A0A2M4BT52 A0A2M4BU94 A0A2M4BTQ9 Q5TXG6 A0A182K935 A0A182FR47 Q7PCV5 A0A1B6D1X3 A0A0K8TS26 A0A182IXE1 A0A182KXT1 A0A182WVK0 A0A182VPA4 A0A1I8NRX4 A0A182PZY3 A0A182I978 A0A3B0JXU8 A0A1B6DQD8 A0A3B0K549 A0A2M4CY36 A0A2M4AHF5 A0A2M4AHG0 A0A2M4AHK1 A0A2M4AHG8 A0A0N7ZCB0 A0A2M4CXQ8 B4L6Z7 A0A0P4WE55 A0A2M4DNH9 A0A1W4VG81 A0A1I8M130 A0A2M4DNE9 A0A084VKD9 A0A1I8M126 A0A1Y1LWE6 A0A1Q3FK33 A0A0L0C132 B4PWR0 B4GWB4 W5JEE7 B3NTI0 A0A0A1X000 A0A1Y1LTU0 A0A034VJZ0 E9IQ80 B5DLL2 B0X9R2 K7J5N1 A0A154PPV7 B4R5F9 A0A0J7JZN3 B4IKA4 B3MZ15 B4NQ41 A0A034VLK8
A0A212EMJ0 A0A2H1WQU5 H9JCL6 A0A194QCS1 A0A2A4JMV9 A0A212FA52 A0A194QX54 D6WX88 A0A1W4WIP4 A0A1W4WUU4 A0A1W4WTJ0 A0A0T6AX11 S4P8F4 A0A2J7PKN1 A0A067R6N0 N6U4S0 U4UAU5 A0A2J7PKL9 A0A182RB96 A0A1J1I3V7 A0A182P406 A0A023ENS3 A0A182M9X4 A0A2M3Z6F6 A0A182SS98 A0A2M3Z695 A0A2M3Z627 A0A2M3Z5Y8 A0A3R7MJ34 A0A182H9K5 A0A182YG36 A0A182W106 A0A182ND52 A0A336K395 T1DR80 U5EQK4 A0A2M4BTW3 A0A2M4BTC5 A0A2M4BT52 A0A2M4BU94 A0A2M4BTQ9 Q5TXG6 A0A182K935 A0A182FR47 Q7PCV5 A0A1B6D1X3 A0A0K8TS26 A0A182IXE1 A0A182KXT1 A0A182WVK0 A0A182VPA4 A0A1I8NRX4 A0A182PZY3 A0A182I978 A0A3B0JXU8 A0A1B6DQD8 A0A3B0K549 A0A2M4CY36 A0A2M4AHF5 A0A2M4AHG0 A0A2M4AHK1 A0A2M4AHG8 A0A0N7ZCB0 A0A2M4CXQ8 B4L6Z7 A0A0P4WE55 A0A2M4DNH9 A0A1W4VG81 A0A1I8M130 A0A2M4DNE9 A0A084VKD9 A0A1I8M126 A0A1Y1LWE6 A0A1Q3FK33 A0A0L0C132 B4PWR0 B4GWB4 W5JEE7 B3NTI0 A0A0A1X000 A0A1Y1LTU0 A0A034VJZ0 E9IQ80 B5DLL2 B0X9R2 K7J5N1 A0A154PPV7 B4R5F9 A0A0J7JZN3 B4IKA4 B3MZ15 B4NQ41 A0A034VLK8
Pubmed
EMBL
NWSH01001074
PCG72744.1
BABH01034706
RSAL01000006
RVE54196.1
ODYU01010378
+ More
SOQ55450.1 KQ459232 KPJ02781.1 KQ461155 KPJ08155.1 AGBW02013813 OWR42705.1 SOQ55451.1 BABH01034705 KPJ02780.1 PCG72742.1 PCG72743.1 AGBW02009515 OWR50616.1 KPJ08156.1 KQ971361 EFA08806.2 LJIG01022605 KRT79676.1 GAIX01005961 JAA86599.1 NEVH01024535 PNF16885.1 KK852663 KDR19066.1 APGK01040155 APGK01040156 KB740975 ENN76585.1 KB632267 ERL91034.1 PNF16883.1 CVRI01000040 CRK94992.1 GAPW01003007 JAC10591.1 AXCM01000042 GGFM01003361 MBW24112.1 GGFM01003279 MBW24030.1 GGFM01003147 MBW23898.1 GGFM01003107 MBW23858.1 QCYY01000609 ROT84025.1 JXUM01121026 JXUM01121027 KQ566454 KXJ70125.1 UFQS01000042 UFQT01000042 SSW98332.1 SSX18718.1 GAMD01000760 JAB00831.1 GANO01004202 JAB55669.1 GGFJ01007301 MBW56442.1 GGFJ01007186 MBW56327.1 GGFJ01007114 MBW56255.1 GGFJ01007187 MBW56328.1 GGFJ01007190 MBW56331.1 AAAB01008592 EAA03450.5 AAAB01008200 EAA03301.4 GEDC01017652 JAS19646.1 GDAI01000650 JAI16953.1 AXCN02001602 APCN01003217 OUUW01000003 SPP78589.1 GEDC01009394 JAS27904.1 SPP78588.1 GGFL01006068 MBW70246.1 GGFK01006889 MBW40210.1 GGFK01006902 MBW40223.1 GGFK01006948 MBW40269.1 GGFK01006915 MBW40236.1 GDRN01070068 JAI63944.1 GGFL01005938 MBW70116.1 CH933812 EDW06143.2 KRG07162.1 GDRN01070067 JAI63945.1 GGFL01014903 MBW79081.1 GGFL01014904 MBW79082.1 ATLV01014180 KE524947 KFB38433.1 GEZM01050849 JAV75347.1 GFDL01007147 JAV27898.1 JRES01001143 KNC25169.1 CM000162 EDX01806.1 CH479194 EDW26998.1 ADMH02001628 ETN61723.1 CH954180 EDV46984.1 GBXI01009653 JAD04639.1 GEZM01050859 JAV75335.1 GAKP01016515 JAC42437.1 GL764666 EFZ17267.1 CH379063 EDY72414.1 KRT06283.1 DS232546 EDS43264.1 KQ435022 KZC13923.1 CM000366 EDX18004.1 LBMM01019544 KMQ83514.1 CH480852 EDW51496.1 CH902632 EDV32859.1 CH964291 EDW86266.1 GAKP01016514 JAC42438.1
SOQ55450.1 KQ459232 KPJ02781.1 KQ461155 KPJ08155.1 AGBW02013813 OWR42705.1 SOQ55451.1 BABH01034705 KPJ02780.1 PCG72742.1 PCG72743.1 AGBW02009515 OWR50616.1 KPJ08156.1 KQ971361 EFA08806.2 LJIG01022605 KRT79676.1 GAIX01005961 JAA86599.1 NEVH01024535 PNF16885.1 KK852663 KDR19066.1 APGK01040155 APGK01040156 KB740975 ENN76585.1 KB632267 ERL91034.1 PNF16883.1 CVRI01000040 CRK94992.1 GAPW01003007 JAC10591.1 AXCM01000042 GGFM01003361 MBW24112.1 GGFM01003279 MBW24030.1 GGFM01003147 MBW23898.1 GGFM01003107 MBW23858.1 QCYY01000609 ROT84025.1 JXUM01121026 JXUM01121027 KQ566454 KXJ70125.1 UFQS01000042 UFQT01000042 SSW98332.1 SSX18718.1 GAMD01000760 JAB00831.1 GANO01004202 JAB55669.1 GGFJ01007301 MBW56442.1 GGFJ01007186 MBW56327.1 GGFJ01007114 MBW56255.1 GGFJ01007187 MBW56328.1 GGFJ01007190 MBW56331.1 AAAB01008592 EAA03450.5 AAAB01008200 EAA03301.4 GEDC01017652 JAS19646.1 GDAI01000650 JAI16953.1 AXCN02001602 APCN01003217 OUUW01000003 SPP78589.1 GEDC01009394 JAS27904.1 SPP78588.1 GGFL01006068 MBW70246.1 GGFK01006889 MBW40210.1 GGFK01006902 MBW40223.1 GGFK01006948 MBW40269.1 GGFK01006915 MBW40236.1 GDRN01070068 JAI63944.1 GGFL01005938 MBW70116.1 CH933812 EDW06143.2 KRG07162.1 GDRN01070067 JAI63945.1 GGFL01014903 MBW79081.1 GGFL01014904 MBW79082.1 ATLV01014180 KE524947 KFB38433.1 GEZM01050849 JAV75347.1 GFDL01007147 JAV27898.1 JRES01001143 KNC25169.1 CM000162 EDX01806.1 CH479194 EDW26998.1 ADMH02001628 ETN61723.1 CH954180 EDV46984.1 GBXI01009653 JAD04639.1 GEZM01050859 JAV75335.1 GAKP01016515 JAC42437.1 GL764666 EFZ17267.1 CH379063 EDY72414.1 KRT06283.1 DS232546 EDS43264.1 KQ435022 KZC13923.1 CM000366 EDX18004.1 LBMM01019544 KMQ83514.1 CH480852 EDW51496.1 CH902632 EDV32859.1 CH964291 EDW86266.1 GAKP01016514 JAC42438.1
Proteomes
UP000218220
UP000005204
UP000283053
UP000053268
UP000053240
UP000007151
+ More
UP000007266 UP000192223 UP000235965 UP000027135 UP000019118 UP000030742 UP000075900 UP000183832 UP000075885 UP000075883 UP000075901 UP000283509 UP000069940 UP000249989 UP000076408 UP000075920 UP000075884 UP000007062 UP000075881 UP000069272 UP000075880 UP000075882 UP000076407 UP000075903 UP000095300 UP000075886 UP000075840 UP000268350 UP000009192 UP000192221 UP000095301 UP000030765 UP000037069 UP000002282 UP000008744 UP000000673 UP000008711 UP000001819 UP000002320 UP000002358 UP000076502 UP000000304 UP000036403 UP000001292 UP000007801 UP000007798
UP000007266 UP000192223 UP000235965 UP000027135 UP000019118 UP000030742 UP000075900 UP000183832 UP000075885 UP000075883 UP000075901 UP000283509 UP000069940 UP000249989 UP000076408 UP000075920 UP000075884 UP000007062 UP000075881 UP000069272 UP000075880 UP000075882 UP000076407 UP000075903 UP000095300 UP000075886 UP000075840 UP000268350 UP000009192 UP000192221 UP000095301 UP000030765 UP000037069 UP000002282 UP000008744 UP000000673 UP000008711 UP000001819 UP000002320 UP000002358 UP000076502 UP000000304 UP000036403 UP000001292 UP000007801 UP000007798
Interpro
SUPFAM
SSF55811
SSF55811
ProteinModelPortal
A0A2A4JMD5
H9JCQ4
A0A3S2P0X8
A0A2H1WQU9
A0A194QH61
A0A194QT27
+ More
A0A212EMJ0 A0A2H1WQU5 H9JCL6 A0A194QCS1 A0A2A4JMV9 A0A212FA52 A0A194QX54 D6WX88 A0A1W4WIP4 A0A1W4WUU4 A0A1W4WTJ0 A0A0T6AX11 S4P8F4 A0A2J7PKN1 A0A067R6N0 N6U4S0 U4UAU5 A0A2J7PKL9 A0A182RB96 A0A1J1I3V7 A0A182P406 A0A023ENS3 A0A182M9X4 A0A2M3Z6F6 A0A182SS98 A0A2M3Z695 A0A2M3Z627 A0A2M3Z5Y8 A0A3R7MJ34 A0A182H9K5 A0A182YG36 A0A182W106 A0A182ND52 A0A336K395 T1DR80 U5EQK4 A0A2M4BTW3 A0A2M4BTC5 A0A2M4BT52 A0A2M4BU94 A0A2M4BTQ9 Q5TXG6 A0A182K935 A0A182FR47 Q7PCV5 A0A1B6D1X3 A0A0K8TS26 A0A182IXE1 A0A182KXT1 A0A182WVK0 A0A182VPA4 A0A1I8NRX4 A0A182PZY3 A0A182I978 A0A3B0JXU8 A0A1B6DQD8 A0A3B0K549 A0A2M4CY36 A0A2M4AHF5 A0A2M4AHG0 A0A2M4AHK1 A0A2M4AHG8 A0A0N7ZCB0 A0A2M4CXQ8 B4L6Z7 A0A0P4WE55 A0A2M4DNH9 A0A1W4VG81 A0A1I8M130 A0A2M4DNE9 A0A084VKD9 A0A1I8M126 A0A1Y1LWE6 A0A1Q3FK33 A0A0L0C132 B4PWR0 B4GWB4 W5JEE7 B3NTI0 A0A0A1X000 A0A1Y1LTU0 A0A034VJZ0 E9IQ80 B5DLL2 B0X9R2 K7J5N1 A0A154PPV7 B4R5F9 A0A0J7JZN3 B4IKA4 B3MZ15 B4NQ41 A0A034VLK8
A0A212EMJ0 A0A2H1WQU5 H9JCL6 A0A194QCS1 A0A2A4JMV9 A0A212FA52 A0A194QX54 D6WX88 A0A1W4WIP4 A0A1W4WUU4 A0A1W4WTJ0 A0A0T6AX11 S4P8F4 A0A2J7PKN1 A0A067R6N0 N6U4S0 U4UAU5 A0A2J7PKL9 A0A182RB96 A0A1J1I3V7 A0A182P406 A0A023ENS3 A0A182M9X4 A0A2M3Z6F6 A0A182SS98 A0A2M3Z695 A0A2M3Z627 A0A2M3Z5Y8 A0A3R7MJ34 A0A182H9K5 A0A182YG36 A0A182W106 A0A182ND52 A0A336K395 T1DR80 U5EQK4 A0A2M4BTW3 A0A2M4BTC5 A0A2M4BT52 A0A2M4BU94 A0A2M4BTQ9 Q5TXG6 A0A182K935 A0A182FR47 Q7PCV5 A0A1B6D1X3 A0A0K8TS26 A0A182IXE1 A0A182KXT1 A0A182WVK0 A0A182VPA4 A0A1I8NRX4 A0A182PZY3 A0A182I978 A0A3B0JXU8 A0A1B6DQD8 A0A3B0K549 A0A2M4CY36 A0A2M4AHF5 A0A2M4AHG0 A0A2M4AHK1 A0A2M4AHG8 A0A0N7ZCB0 A0A2M4CXQ8 B4L6Z7 A0A0P4WE55 A0A2M4DNH9 A0A1W4VG81 A0A1I8M130 A0A2M4DNE9 A0A084VKD9 A0A1I8M126 A0A1Y1LWE6 A0A1Q3FK33 A0A0L0C132 B4PWR0 B4GWB4 W5JEE7 B3NTI0 A0A0A1X000 A0A1Y1LTU0 A0A034VJZ0 E9IQ80 B5DLL2 B0X9R2 K7J5N1 A0A154PPV7 B4R5F9 A0A0J7JZN3 B4IKA4 B3MZ15 B4NQ41 A0A034VLK8
PDB
4ZBP
E-value=4.07077e-28,
Score=308
Ontologies
GO
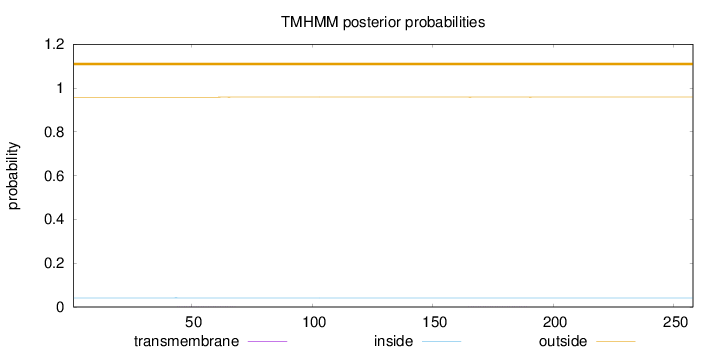
Topology
Length:
258
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02181
Exp number, first 60 AAs:
0.00853
Total prob of N-in:
0.04159
outside
1 - 258
Population Genetic Test Statistics
Pi
202.345502
Theta
52.683052
Tajima's D
0.136808
CLR
0.654788
CSRT
0.401029948502575
Interpretation
Uncertain
