Pre Gene Modal
BGIBMGA007299
Annotation
PREDICTED:_Golgi_to_ER_traffic_protein_4_homolog_[Amyelois_transitella]
Full name
Golgi to ER traffic protein 4 homolog
Alternative Name
Conserved edge expressed protein
Location in the cell
Cytoplasmic Reliability : 1.156 PlasmaMembrane Reliability : 1.299
Sequence
CDS
ATGGCCACGCGTGGAGAAAGAGGAGTTGTTAGGGTATTAGATAAGTTAGAAGCTTCAATTAATTCTGGCCAGTATTATGAAGCACATCAAATGTACAGAACTCTTTATTTTAGATACCTAACTCAGAAAAAATATCCAGAATTGTTAACTTTATTGTACAAAGGGTCTACACTTCTTTTGGAACGCGATCAACAAGGAAGTGGAGCTGATCTCGCCATACTCTTCGTAGAAGTTTTAACAAAATCGGAAACCAAACCTAATGAAGAATGGGTCTCGAAATTGGCAAAATTATTTGAATTAATCAGCTCCAGTGTCCCAGAGCGAGAAACCTTTCTAACAAATTCAATAAAATGGTCAATGGATAACAACAAAAAAGGTGACCCACTCCTACACAAGAAAATAGCAGAAGTATTTTGGAAGGAAAAAAAGTACACAGCTGCTCATAGACACTTTTTACACTCATCGGATGGTTCAGCATATGCTAACATGCTTATAGAGTTACACACAACTAAAGGTCTCAAATCAGAAATAGATTTGTTCATAGCACAAGCTGTGTTGCAAATACTTTGTTTGAGAAATGTAAGAATGGCAACTGAAACATTTTTGAGGTATACAGAAACACATCCCAAAATTAAGAATAGCAAAGGACCTCCATATATATTCCCACTATTGAATTTTTTGTGGTTCTTGCTGAGAGCTGTCGAACAGAGACATGTACGACAGTTTATGGTACTAAAGAATTGGTATGCAATTAGCATTAAAAGGGATCCAAACTACTCCATTTTTTTAGACAATATAGGCCGCATTTGGTTTGGAATAGAAATACCTTCGGAAAATAGAAATCCAAACTCTATGTTCGGAGGTCTAATAAAATCTATTATTGGTGATATAGATAGTTCTGATGATGATGGTGAAAGCAAGACGTCGGCCACAGCTCCTGATTTGGACTAA
Protein
MATRGERGVVRVLDKLEASINSGQYYEAHQMYRTLYFRYLTQKKYPELLTLLYKGSTLLLERDQQGSGADLAILFVEVLTKSETKPNEEWVSKLAKLFELISSSVPERETFLTNSIKWSMDNNKKGDPLLHKKIAEVFWKEKKYTAAHRHFLHSSDGSAYANMLIELHTTKGLKSEIDLFIAQAVLQILCLRNVRMATETFLRYTETHPKIKNSKGPPYIFPLLNFLWFLLRAVEQRHVRQFMVLKNWYAISIKRDPNYSIFLDNIGRIWFGIEIPSENRNPNSMFGGLIKSIIGDIDSSDDDGESKTSATAPDLD
Summary
Description
As part of a cytosolic protein quality control complex, the bag6/bat3 complex, maintains misfolded and hydrophobic patches-containing proteins in a soluble state and participates to their proper delivery to the endoplasmic reticulum or alternatively can promote their sorting to the proteasome where they undergo degradation. The bag6/bat3 complex is involved in the post-translational delivery of tail-anchored/type II transmembrane proteins to the endoplasmic reticulum membrane. Similarly, the bag6/bat3 complex also functions as a sorting platform for proteins of the secretory pathway that are mislocalized to the cytosol either delivering them to the proteasome for degradation or to the endoplasmic reticulum. The bag6/bat3 complex also plays a role in the endoplasmic reticulum-associated degradation (ERAD), a quality control mechanism that eliminates unwanted proteins of the endoplasmic reticulum through their retrotranslocation to the cytosol and their targeting to the proteasome. It maintains these retrotranslocated proteins in an unfolded yet soluble state condition in the cytosol to ensure their proper delivery to the proteasome.
Subunit
Component of the bag6/bat3 complex.
Similarity
Belongs to the GET4 family.
Keywords
Complete proteome
Cytoplasm
Reference proteome
Transport
Feature
chain Golgi to ER traffic protein 4 homolog
Uniprot
H9JCQ2
A0A3S2M8X6
A0A1E1WNR8
A0A2A4JL93
A0A2H1WR60
A0A212FA41
+ More
S4PPR3 A0A194QRP1 A0A0L7KZL7 A0A1W4WIC0 A0A224XFM0 A0A069DRS9 A0A1B6C9S1 A0A2J7PLV0 A0A0V0G5B3 D6WYB6 A0A0N7Z8W5 V5GRX4 A0A1Y1KLU4 A0A067RMM6 A0A1B6GIM5 J3JZ83 A0A1B6MMB8 A0A0A9XXR2 A0A0T6B1V3 A0A2R5LL13 A0A023GKP0 E0VFK7 A0A023FUM9 A0A3B4X9H1 A0A3B4TVK6 A0A146WB57 A0A2I4CSJ7 A0A0F8AEY0 A0A3B3TJV2 A0A3B3WIU1 A0A3Q2QTI7 A0A3Q2C901 A0A3Q0T5K8 A0A147A2R7 A0A087YH40 E6ZGL2 A0A315VYD5 A0A3Q3JIB5 G3NMJ4 A0A023EQ78 A0A182GL78 A0A3Q1HNU9 A0A1E1XBY1 A0A3Q3KWN9 A0A3Q3QJ17 A0A3P9QAK0 A0A3P9J0P2 M4A5E0 A0A0S7G0A1 A0A3Q1GHZ0 A0A3B5BJY7 A0A3P9M0C1 H2N1E0 A0A3Q2XS96 A0A3Q1AKD7 A0A1A7YBN2 A0A1A7XI13 A0A0S7FSZ8 A0A3Q2W7R7 A0A182H6W4 A0A3R7LYG1 A0A3B4F2U3 A0A3Q2X8N8 A0A3Q4GEU8 I3KUM4 E2B1Q9 A0A1A8CAF3 A0A154PPY3 A0A3P8WYI3 A4QNE0 A0A0N8K2W0 A0A226NJT4 A0A226PTK4 G1K3K2 A0A1A8DKN6 A0A1A8JXI0 A0A1A8MFQ4 A0A1A8NLH0 A0A1A8RKJ7 A0A1A8IX59 E2BIS1 A0A3P8TYM2 Q16KN0 G3NMK3 A1Z3X3 A0A1A8PN16 A0A287AWN6 A0A1A8UBT3 A0A1A7ZXW2 A0A2Y9RVL8 A0A3B3BSQ4 H2V3I3 A0A0P4WP96
S4PPR3 A0A194QRP1 A0A0L7KZL7 A0A1W4WIC0 A0A224XFM0 A0A069DRS9 A0A1B6C9S1 A0A2J7PLV0 A0A0V0G5B3 D6WYB6 A0A0N7Z8W5 V5GRX4 A0A1Y1KLU4 A0A067RMM6 A0A1B6GIM5 J3JZ83 A0A1B6MMB8 A0A0A9XXR2 A0A0T6B1V3 A0A2R5LL13 A0A023GKP0 E0VFK7 A0A023FUM9 A0A3B4X9H1 A0A3B4TVK6 A0A146WB57 A0A2I4CSJ7 A0A0F8AEY0 A0A3B3TJV2 A0A3B3WIU1 A0A3Q2QTI7 A0A3Q2C901 A0A3Q0T5K8 A0A147A2R7 A0A087YH40 E6ZGL2 A0A315VYD5 A0A3Q3JIB5 G3NMJ4 A0A023EQ78 A0A182GL78 A0A3Q1HNU9 A0A1E1XBY1 A0A3Q3KWN9 A0A3Q3QJ17 A0A3P9QAK0 A0A3P9J0P2 M4A5E0 A0A0S7G0A1 A0A3Q1GHZ0 A0A3B5BJY7 A0A3P9M0C1 H2N1E0 A0A3Q2XS96 A0A3Q1AKD7 A0A1A7YBN2 A0A1A7XI13 A0A0S7FSZ8 A0A3Q2W7R7 A0A182H6W4 A0A3R7LYG1 A0A3B4F2U3 A0A3Q2X8N8 A0A3Q4GEU8 I3KUM4 E2B1Q9 A0A1A8CAF3 A0A154PPY3 A0A3P8WYI3 A4QNE0 A0A0N8K2W0 A0A226NJT4 A0A226PTK4 G1K3K2 A0A1A8DKN6 A0A1A8JXI0 A0A1A8MFQ4 A0A1A8NLH0 A0A1A8RKJ7 A0A1A8IX59 E2BIS1 A0A3P8TYM2 Q16KN0 G3NMK3 A1Z3X3 A0A1A8PN16 A0A287AWN6 A0A1A8UBT3 A0A1A7ZXW2 A0A2Y9RVL8 A0A3B3BSQ4 H2V3I3 A0A0P4WP96
Pubmed
EMBL
BABH01034704
RSAL01000006
RVE54193.1
GDQN01002456
JAT88598.1
NWSH01001074
+ More
PCG72741.1 ODYU01010378 SOQ55452.1 AGBW02009515 OWR50615.1 GAIX01000077 JAA92483.1 KQ461155 KPJ08157.1 JTDY01004024 KOB68698.1 GFTR01005149 JAW11277.1 GBGD01002111 JAC86778.1 GEDC01027097 JAS10201.1 NEVH01024423 PNF17303.1 GECL01002876 JAP03248.1 KQ971356 EFA09129.1 GDKW01002211 JAI54384.1 GALX01005493 JAB62973.1 GEZM01079835 JAV62373.1 KK852421 KDR24283.1 GECZ01007485 JAS62284.1 BT128564 AEE63521.1 GEBQ01002915 JAT37062.1 GBHO01019966 GDHC01004584 JAG23638.1 JAQ14045.1 LJIG01016292 KRT81038.1 GGLE01005881 MBY10007.1 GBBM01001740 JAC33678.1 DS235115 EEB12163.1 GBBK01000102 JAC24380.1 GCES01059015 JAR27308.1 KQ042663 KKF12255.1 GCES01013838 JAR72485.1 AYCK01000769 FQ310507 CBN81196.1 NHOQ01000843 PWA28360.1 GAPW01002365 JAC11233.1 JXUM01071389 KQ562662 KXJ75372.1 GFAC01002421 JAT96767.1 GBYX01461421 JAO20147.1 HADX01005685 SBP27917.1 HADW01016243 SBP17643.1 GBYX01461433 JAO20135.1 JXUM01114983 KQ565837 KXJ70615.1 QCYY01002914 ROT66639.1 AERX01002224 GL444953 EFN60415.1 HADZ01011820 SBP75761.1 KQ435012 KZC13777.1 BC135489 JARO02000366 KPP78833.1 MCFN01000035 OXB67598.1 AWGT02000022 OXB82727.1 AAMC01119009 HAEA01005346 SBQ33826.1 HAEE01004627 SBR24647.1 HAEF01014045 SBR55204.1 HAEG01003732 SBR69841.1 HAEI01009027 SBS06635.1 HAED01015504 SBR01949.1 GL448530 EFN84411.1 CH477950 CH477244 EAT34855.1 EAT46261.1 EF177382 HAEH01007639 SBR82407.1 AEMK02000013 DQIR01215747 DQIR01225601 DQIR01307325 HDB71224.1 HDC62803.1 HAEJ01004126 SBS44583.1 HADY01008626 SBP47111.1 GDRN01074368 JAI63254.1
PCG72741.1 ODYU01010378 SOQ55452.1 AGBW02009515 OWR50615.1 GAIX01000077 JAA92483.1 KQ461155 KPJ08157.1 JTDY01004024 KOB68698.1 GFTR01005149 JAW11277.1 GBGD01002111 JAC86778.1 GEDC01027097 JAS10201.1 NEVH01024423 PNF17303.1 GECL01002876 JAP03248.1 KQ971356 EFA09129.1 GDKW01002211 JAI54384.1 GALX01005493 JAB62973.1 GEZM01079835 JAV62373.1 KK852421 KDR24283.1 GECZ01007485 JAS62284.1 BT128564 AEE63521.1 GEBQ01002915 JAT37062.1 GBHO01019966 GDHC01004584 JAG23638.1 JAQ14045.1 LJIG01016292 KRT81038.1 GGLE01005881 MBY10007.1 GBBM01001740 JAC33678.1 DS235115 EEB12163.1 GBBK01000102 JAC24380.1 GCES01059015 JAR27308.1 KQ042663 KKF12255.1 GCES01013838 JAR72485.1 AYCK01000769 FQ310507 CBN81196.1 NHOQ01000843 PWA28360.1 GAPW01002365 JAC11233.1 JXUM01071389 KQ562662 KXJ75372.1 GFAC01002421 JAT96767.1 GBYX01461421 JAO20147.1 HADX01005685 SBP27917.1 HADW01016243 SBP17643.1 GBYX01461433 JAO20135.1 JXUM01114983 KQ565837 KXJ70615.1 QCYY01002914 ROT66639.1 AERX01002224 GL444953 EFN60415.1 HADZ01011820 SBP75761.1 KQ435012 KZC13777.1 BC135489 JARO02000366 KPP78833.1 MCFN01000035 OXB67598.1 AWGT02000022 OXB82727.1 AAMC01119009 HAEA01005346 SBQ33826.1 HAEE01004627 SBR24647.1 HAEF01014045 SBR55204.1 HAEG01003732 SBR69841.1 HAEI01009027 SBS06635.1 HAED01015504 SBR01949.1 GL448530 EFN84411.1 CH477950 CH477244 EAT34855.1 EAT46261.1 EF177382 HAEH01007639 SBR82407.1 AEMK02000013 DQIR01215747 DQIR01225601 DQIR01307325 HDB71224.1 HDC62803.1 HAEJ01004126 SBS44583.1 HADY01008626 SBP47111.1 GDRN01074368 JAI63254.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000007151
UP000053240
UP000037510
+ More
UP000192223 UP000235965 UP000007266 UP000027135 UP000009046 UP000261360 UP000261420 UP000192220 UP000261500 UP000261480 UP000265000 UP000265020 UP000261340 UP000028760 UP000261600 UP000007635 UP000069940 UP000249989 UP000265040 UP000261660 UP000242638 UP000265200 UP000002852 UP000257200 UP000261400 UP000265180 UP000001038 UP000264820 UP000257160 UP000264840 UP000283509 UP000261460 UP000261580 UP000005207 UP000000311 UP000076502 UP000265120 UP000008143 UP000034805 UP000192224 UP000198323 UP000198419 UP000008237 UP000265080 UP000008820 UP000008227 UP000248480 UP000261560 UP000005226
UP000192223 UP000235965 UP000007266 UP000027135 UP000009046 UP000261360 UP000261420 UP000192220 UP000261500 UP000261480 UP000265000 UP000265020 UP000261340 UP000028760 UP000261600 UP000007635 UP000069940 UP000249989 UP000265040 UP000261660 UP000242638 UP000265200 UP000002852 UP000257200 UP000261400 UP000265180 UP000001038 UP000264820 UP000257160 UP000264840 UP000283509 UP000261460 UP000261580 UP000005207 UP000000311 UP000076502 UP000265120 UP000008143 UP000034805 UP000192224 UP000198323 UP000198419 UP000008237 UP000265080 UP000008820 UP000008227 UP000248480 UP000261560 UP000005226
PRIDE
Pfam
PF04190 DUF410
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
H9JCQ2
A0A3S2M8X6
A0A1E1WNR8
A0A2A4JL93
A0A2H1WR60
A0A212FA41
+ More
S4PPR3 A0A194QRP1 A0A0L7KZL7 A0A1W4WIC0 A0A224XFM0 A0A069DRS9 A0A1B6C9S1 A0A2J7PLV0 A0A0V0G5B3 D6WYB6 A0A0N7Z8W5 V5GRX4 A0A1Y1KLU4 A0A067RMM6 A0A1B6GIM5 J3JZ83 A0A1B6MMB8 A0A0A9XXR2 A0A0T6B1V3 A0A2R5LL13 A0A023GKP0 E0VFK7 A0A023FUM9 A0A3B4X9H1 A0A3B4TVK6 A0A146WB57 A0A2I4CSJ7 A0A0F8AEY0 A0A3B3TJV2 A0A3B3WIU1 A0A3Q2QTI7 A0A3Q2C901 A0A3Q0T5K8 A0A147A2R7 A0A087YH40 E6ZGL2 A0A315VYD5 A0A3Q3JIB5 G3NMJ4 A0A023EQ78 A0A182GL78 A0A3Q1HNU9 A0A1E1XBY1 A0A3Q3KWN9 A0A3Q3QJ17 A0A3P9QAK0 A0A3P9J0P2 M4A5E0 A0A0S7G0A1 A0A3Q1GHZ0 A0A3B5BJY7 A0A3P9M0C1 H2N1E0 A0A3Q2XS96 A0A3Q1AKD7 A0A1A7YBN2 A0A1A7XI13 A0A0S7FSZ8 A0A3Q2W7R7 A0A182H6W4 A0A3R7LYG1 A0A3B4F2U3 A0A3Q2X8N8 A0A3Q4GEU8 I3KUM4 E2B1Q9 A0A1A8CAF3 A0A154PPY3 A0A3P8WYI3 A4QNE0 A0A0N8K2W0 A0A226NJT4 A0A226PTK4 G1K3K2 A0A1A8DKN6 A0A1A8JXI0 A0A1A8MFQ4 A0A1A8NLH0 A0A1A8RKJ7 A0A1A8IX59 E2BIS1 A0A3P8TYM2 Q16KN0 G3NMK3 A1Z3X3 A0A1A8PN16 A0A287AWN6 A0A1A8UBT3 A0A1A7ZXW2 A0A2Y9RVL8 A0A3B3BSQ4 H2V3I3 A0A0P4WP96
S4PPR3 A0A194QRP1 A0A0L7KZL7 A0A1W4WIC0 A0A224XFM0 A0A069DRS9 A0A1B6C9S1 A0A2J7PLV0 A0A0V0G5B3 D6WYB6 A0A0N7Z8W5 V5GRX4 A0A1Y1KLU4 A0A067RMM6 A0A1B6GIM5 J3JZ83 A0A1B6MMB8 A0A0A9XXR2 A0A0T6B1V3 A0A2R5LL13 A0A023GKP0 E0VFK7 A0A023FUM9 A0A3B4X9H1 A0A3B4TVK6 A0A146WB57 A0A2I4CSJ7 A0A0F8AEY0 A0A3B3TJV2 A0A3B3WIU1 A0A3Q2QTI7 A0A3Q2C901 A0A3Q0T5K8 A0A147A2R7 A0A087YH40 E6ZGL2 A0A315VYD5 A0A3Q3JIB5 G3NMJ4 A0A023EQ78 A0A182GL78 A0A3Q1HNU9 A0A1E1XBY1 A0A3Q3KWN9 A0A3Q3QJ17 A0A3P9QAK0 A0A3P9J0P2 M4A5E0 A0A0S7G0A1 A0A3Q1GHZ0 A0A3B5BJY7 A0A3P9M0C1 H2N1E0 A0A3Q2XS96 A0A3Q1AKD7 A0A1A7YBN2 A0A1A7XI13 A0A0S7FSZ8 A0A3Q2W7R7 A0A182H6W4 A0A3R7LYG1 A0A3B4F2U3 A0A3Q2X8N8 A0A3Q4GEU8 I3KUM4 E2B1Q9 A0A1A8CAF3 A0A154PPY3 A0A3P8WYI3 A4QNE0 A0A0N8K2W0 A0A226NJT4 A0A226PTK4 G1K3K2 A0A1A8DKN6 A0A1A8JXI0 A0A1A8MFQ4 A0A1A8NLH0 A0A1A8RKJ7 A0A1A8IX59 E2BIS1 A0A3P8TYM2 Q16KN0 G3NMK3 A1Z3X3 A0A1A8PN16 A0A287AWN6 A0A1A8UBT3 A0A1A7ZXW2 A0A2Y9RVL8 A0A3B3BSQ4 H2V3I3 A0A0P4WP96
PDB
6AU8
E-value=8.10771e-67,
Score=643
Ontologies
KEGG
GO
PANTHER
Topology
Subcellular location
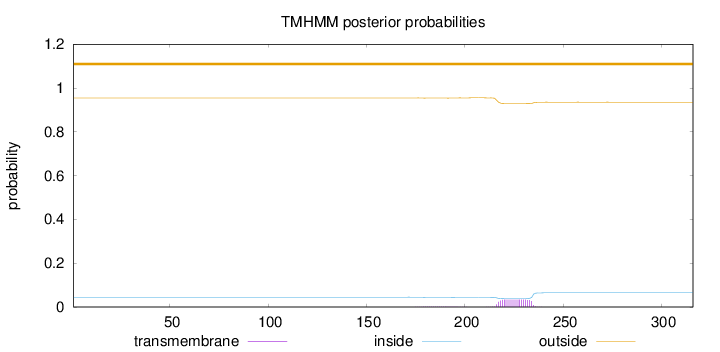
Length:
316
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.648810000000001
Exp number, first 60 AAs:
0.00077
Total prob of N-in:
0.04501
outside
1 - 316
Population Genetic Test Statistics
Pi
219.207903
Theta
160.181601
Tajima's D
0.739619
CLR
0.430069
CSRT
0.587170641467927
Interpretation
Uncertain
