Pre Gene Modal
BGIBMGA007265
Annotation
PREDICTED:_translation_factor_GUF1_homolog?_mitochondrial_isoform_X1_[Papilio_machaon]
Full name
Translation factor GUF1 homolog, mitochondrial
+ More
Translation factor waclaw, mitochondrial
Translation factor GUF1, mitochondrial
Translation factor waclaw, mitochondrial
Translation factor GUF1, mitochondrial
Alternative Name
Elongation factor 4 homolog
GTPase GUF1 homolog
Ribosomal back-translocase
GTPase GUF1
GTPase GUF1 homolog
Ribosomal back-translocase
GTPase GUF1
Location in the cell
Cytoplasmic Reliability : 1.838 Mitochondrial Reliability : 1.474
Sequence
CDS
ATGGTTACTAATATAAATTTTATAAAATACTTGTATTACAATTCTGTTAAATATCATCAAAGGGTCTTAATGAGACGTATGAGTTCAGACTCTAGGAAATATGTAGATGTGGAAAAAATAAGGAACTTTAGCATTGTAGCTCACGTCGATCATGGCAAGAGCACTTTGGCTGACAGACTTCTTGAGTTAACCGGTGTAAAGAAACCGGGCGATGAACAATCTCAAGTTTTGGACCAACTACAGGTTGAAAAAGAAAGAGGAATTACAGTGAAGGCAGTCACGGCTACTCTAGACTACACATATAACAATGAGACGTACCTACTCAATTTAATTGACACTCCTGGACATGTGGATTTCTCCAATGAAGTAGTGAGAAGTGTAACAGCATGTCAAGGAGTCATTATGCTGGTGGATGCTAATGAAGGTATTCAAGCACAAACAGTAGCTGTTCATTCATTAGCTAAGAGAAACAATTTAGTAATAATACCAACACTAAACAAAGTGGATCTTCCAAGAGCTAACCCTGACAAAGTGAAAGAACAGTTACAATCAGTATTCAAAATAGACCCTAATACAGTTTTAAGCATATCAGCAAAAAAAGGTTGGGGTGTGCGAGAGCTACTAGAAGCTGTAATAATGAGGATCCCACCACCAAAAGTTGTCTTAGATGATCATTTCCGTGCTCACATAATAGACACATGGCATGATAAATATAGAGGGGTTTTGTGTTTGGCATATATTACTTCAGGGACAGTTAAAGTTGGACAAGCAGTGAAATGGCAATCTGATTCGAAATTACAAACAGTGAAGCACTTGGCACTGTTGAGACCTACTGAGGAGCCCATTGATGAAGCTATTGCAGGTCAAGTTGTAATGGTAGGTTGTGGACCAAAAGGCGGTGGTTCTGTTGGGGATCAATTAGTAGCTAAAGAATTAGCTAATGCTAGTGAAAGAATATCTGCACCTGCAGTGAAGCACATGGTCTACGCTGGTATATTTCCTGCAGACCAATCACAGCATATATTACTGAGTGATGCAATCAAGAAATTAGCTTTAAATGATTCGGCTGTTTCAGTTACGATAGATTCAAGTCCAGCTCTAGGTCAAGGATGGCGTATTGGATTTCTGGGCTTGTTGCACTTAGATGTTTTTACACAACGTTTATTGCAAGAACACAAAGCCGAGGCAATTCTCACAGCACCTTCAGTGCCCTATAAAGTTAAAATAAAAGGAGCCAAACTTATAAAACATTACAAGACTGATGAAATTTTAATCACAAATCCTTTACAATTACCGGACCCAAACAATGTAGTAGAATATTTCGAACCATTTGTTAAAGGTACAATAATTACACCTGTAGAATACATAGGCGCGGTGACATCCCTGTGCATAGACAGACGGGGCACACCCCTACCGTCCAGCGCAGTCGACGACACGATGACCATGTTACAGTTTATCCTACCGCTATCCGAAATCGTCGTCGATTTTCACGATACATTAAAAAGCATAACTTCAGGATTCGCTAGCTTTGATTATGAAGATGACGGCTTTCACTCCAGCTCTTTGGTCCGAATGGATATTTTAATCAATGGTATGCTTGTTGATGAATTATCAACAATAGTGCACGCTAGTCGACTAGAGTATAATGCGCGGAGATTAGCTGAGAAATTAAAAGAAATGATCCCTAGGCAAATGGTGCAGGTTGCCATACAAGCAGTAGTGGGGGGAAAAGTATTCGCACGAGAAACCATCAAAGCCTACAGAAAAGACGTCACCGCTAAATTGTACGGCGGCGACGTTACTCGAAGAAAGAAATTATTAAAGCAACAAGCCGAGGGTAAGAAAAAAATGCGCAGCGTAGCGAACATCAAAATTCCGAGAGATACGTTTATAGACGTGTTGAAAAAGTAG
Protein
MVTNINFIKYLYYNSVKYHQRVLMRRMSSDSRKYVDVEKIRNFSIVAHVDHGKSTLADRLLELTGVKKPGDEQSQVLDQLQVEKERGITVKAVTATLDYTYNNETYLLNLIDTPGHVDFSNEVVRSVTACQGVIMLVDANEGIQAQTVAVHSLAKRNNLVIIPTLNKVDLPRANPDKVKEQLQSVFKIDPNTVLSISAKKGWGVRELLEAVIMRIPPPKVVLDDHFRAHIIDTWHDKYRGVLCLAYITSGTVKVGQAVKWQSDSKLQTVKHLALLRPTEEPIDEAIAGQVVMVGCGPKGGGSVGDQLVAKELANASERISAPAVKHMVYAGIFPADQSQHILLSDAIKKLALNDSAVSVTIDSSPALGQGWRIGFLGLLHLDVFTQRLLQEHKAEAILTAPSVPYKVKIKGAKLIKHYKTDEILITNPLQLPDPNNVVEYFEPFVKGTIITPVEYIGAVTSLCIDRRGTPLPSSAVDDTMTMLQFILPLSEIVVDFHDTLKSITSGFASFDYEDDGFHSSSLVRMDILINGMLVDELSTIVHASRLEYNARRLAEKLKEMIPRQMVQVAIQAVVGGKVFARETIKAYRKDVTAKLYGGDVTRRKKLLKQQAEGKKKMRSVANIKIPRDTFIDVLKK
Summary
Description
Promotes mitochondrial protein synthesis. May act as a fidelity factor of the translation reaction, by catalyzing a one-codon backward translocation of tRNAs on improperly translocated ribosomes. Binds to mitochondrial ribosomes in a GTP-dependent manner.
Catalytic Activity
GTP + H2O = GDP + H(+) + phosphate
Miscellaneous
This protein may be expected to contain an N-terminal transit peptide but none has been predicted.
Similarity
Belongs to the GTP-binding elongation factor family. LepA subfamily.
Belongs to the TRAFAC class translation factor GTPase superfamily. Classic translation factor GTPase family. LepA subfamily.
Belongs to the TRAFAC class translation factor GTPase superfamily. Classic translation factor GTPase family. LepA subfamily.
Keywords
Complete proteome
GTP-binding
Hydrolase
Membrane
Mitochondrion
Mitochondrion inner membrane
Nucleotide-binding
Protein biosynthesis
Reference proteome
Transit peptide
Feature
chain Translation factor GUF1 homolog, mitochondrial
Uniprot
H9JCM0
A0A2H1W0S4
A0A2A4JLQ2
A0A3S2P9K5
A0A194QT37
A0A194QCR0
+ More
A0A212FDE4 B0WXB8 A0A1Q3FCB3 A0A1Y1L517 Q0IFX5 W5JUN6 A0A182GJD8 A0A182T5N1 A0A182XGW6 A0A182RWS6 A0A182KZ19 A0A182PCG3 A0A182KGD9 Q7PMF6 A0A182W8T0 A0A1S4H2L6 B4JNG0 A0A182M5S3 A0A182TJP4 A0A182IZJ7 A0A182HRB1 A0A0M4F967 A0A182UQQ6 A0A182F0T2 B4NCH5 B4L5D7 A0A182NP40 A0A0A1X703 X4YDX6 Q9VRH6 Q86P01 A0A0K8VQR5 A0A182Q138 A0A1I8MJZ7 B4MAZ7 B5DKL9 B4R3R6 B4HD31 B3NYD8 B3MRW0 B4I6H2 B4PZ34 A0A1B0FCK3 A0A1A9YNV1 A0A3B0JCJ5 A0A1B0AEB4 A0A1I8PR26 A0A1B0APL7 W8BL04 A0A182Y6B8 A0A1A9X1G1 A0A1J1I7Z7 A0A1A9UL95 U4UHZ1 A0A026VTH6 A0A034WD04 A0A1B0DJF5 N6TUM5 A0A195FYN7 A0A0P4VQT4 T1HN81 N6TX65 A0A0N8BER3 E9GF27 A0A0C9RII1 A0A0P5QE36 A0A069DVY7 A0A151IYU8 A0A0P4XS62 A0A0P6GQK2 A0A023F4I2 A0A0P5J3B2 A0A0J7NRZ9 E2A9H9 A0A0P4ZJX9 E9JD47 E2BW13 A0A0P5RFK9 A0A0P4WS90 A0A151I492 A0A158NXT4 A0A0N8ABL3 A0A0P5XBV6 H3AGD0 A0A0P6AZP7 F4WLU1 A0A164RWH3 I3KFH1 A0A1S3RR63 A0A060XKC1 H3AGD1 A0A3P9BCX5 A0A310SDA6 A0A2D0RF47
A0A212FDE4 B0WXB8 A0A1Q3FCB3 A0A1Y1L517 Q0IFX5 W5JUN6 A0A182GJD8 A0A182T5N1 A0A182XGW6 A0A182RWS6 A0A182KZ19 A0A182PCG3 A0A182KGD9 Q7PMF6 A0A182W8T0 A0A1S4H2L6 B4JNG0 A0A182M5S3 A0A182TJP4 A0A182IZJ7 A0A182HRB1 A0A0M4F967 A0A182UQQ6 A0A182F0T2 B4NCH5 B4L5D7 A0A182NP40 A0A0A1X703 X4YDX6 Q9VRH6 Q86P01 A0A0K8VQR5 A0A182Q138 A0A1I8MJZ7 B4MAZ7 B5DKL9 B4R3R6 B4HD31 B3NYD8 B3MRW0 B4I6H2 B4PZ34 A0A1B0FCK3 A0A1A9YNV1 A0A3B0JCJ5 A0A1B0AEB4 A0A1I8PR26 A0A1B0APL7 W8BL04 A0A182Y6B8 A0A1A9X1G1 A0A1J1I7Z7 A0A1A9UL95 U4UHZ1 A0A026VTH6 A0A034WD04 A0A1B0DJF5 N6TUM5 A0A195FYN7 A0A0P4VQT4 T1HN81 N6TX65 A0A0N8BER3 E9GF27 A0A0C9RII1 A0A0P5QE36 A0A069DVY7 A0A151IYU8 A0A0P4XS62 A0A0P6GQK2 A0A023F4I2 A0A0P5J3B2 A0A0J7NRZ9 E2A9H9 A0A0P4ZJX9 E9JD47 E2BW13 A0A0P5RFK9 A0A0P4WS90 A0A151I492 A0A158NXT4 A0A0N8ABL3 A0A0P5XBV6 H3AGD0 A0A0P6AZP7 F4WLU1 A0A164RWH3 I3KFH1 A0A1S3RR63 A0A060XKC1 H3AGD1 A0A3P9BCX5 A0A310SDA6 A0A2D0RF47
EC Number
3.6.5.n1
3.6.5.-
3.6.5.-
Pubmed
19121390
26354079
22118469
28004739
17510324
20920257
+ More
23761445 26483478 20966253 12364791 17994087 25830018 9520435 10731132 12537572 25315136 15632085 17550304 24495485 25244985 23537049 24508170 30249741 25348373 27129103 21292972 26334808 25474469 20798317 21282665 21347285 9215903 21719571 25186727 24755649
23761445 26483478 20966253 12364791 17994087 25830018 9520435 10731132 12537572 25315136 15632085 17550304 24495485 25244985 23537049 24508170 30249741 25348373 27129103 21292972 26334808 25474469 20798317 21282665 21347285 9215903 21719571 25186727 24755649
EMBL
BABH01034693
ODYU01005610
SOQ46627.1
NWSH01001074
PCG72749.1
RSAL01000168
+ More
RVE45234.1 KQ461155 KPJ08165.1 KQ459232 KPJ02770.1 AGBW02009056 OWR51785.1 DS232161 GFDL01009848 JAV25197.1 GEZM01068259 JAV67065.1 CH477286 ADMH02000444 ETN66459.1 JXUM01067886 KQ562472 KXJ75822.1 AAAB01008980 EAA13916.4 CH916371 EDV92253.1 AXCM01000552 APCN01001664 CP012528 ALC49037.1 CH964239 EDW82534.1 CH933811 EDW06396.1 GBXI01009457 GBXI01007405 JAD04835.1 JAD06887.1 BT150439 AHV85501.1 AF017777 AE014298 BT003552 AAO39556.1 GDHF01026205 GDHF01023022 GDHF01020170 GDHF01011116 GDHF01010385 GDHF01001301 JAI26109.1 JAI29292.1 JAI32144.1 JAI41198.1 JAI41929.1 JAI51013.1 AXCN02001967 CH940655 EDW66406.2 CH379063 EDY72069.2 CM000366 EDX18534.1 CH479636 EDW35356.1 CH954180 EDV47617.2 CH902622 EDV34515.2 CH480823 EDW56378.1 CM000162 EDX03095.2 CCAG010004527 CCAG010004528 OUUW01000003 SPP77812.1 JXJN01001483 GAMC01012594 GAMC01012590 JAB93961.1 CVRI01000043 CRK96439.1 KB632308 ERL91963.1 KK107941 QOIP01000004 EZA47093.1 RLU23968.1 GAKP01006927 JAC52025.1 AJVK01015784 APGK01051610 KB741197 ENN72980.1 KQ981204 KYN44944.1 GDKW01001741 JAI54854.1 ACPB03015116 ENN72981.1 GDIQ01184955 JAK66770.1 GL732541 EFX81974.1 GBYB01008055 JAG77822.1 GDIQ01136766 JAL14960.1 GBGD01000893 JAC87996.1 KQ980761 KYN13602.1 GDIP01238823 JAI84578.1 GDIQ01042559 JAN52178.1 GBBI01002620 JAC16092.1 GDIQ01205261 JAK46464.1 LBMM01002135 KMQ95220.1 GL437912 EFN69903.1 GDIP01211684 JAJ11718.1 GL771866 EFZ09158.1 GL451091 EFN80045.1 GDIQ01101444 JAL50282.1 GDRN01066268 GDRN01066267 GDRN01066266 JAI64571.1 KQ976462 KYM84642.1 ADTU01003520 ADTU01003521 GDIP01163433 JAJ59969.1 GDIP01074452 JAM29263.1 AFYH01140933 AFYH01140934 AFYH01140935 AFYH01140936 GDIP01023074 JAM80641.1 GL888216 EGI64823.1 LRGB01002121 KZS09009.1 AERX01026919 AERX01026920 FR905222 CDQ77330.1 KQ761108 OAD58203.1
RVE45234.1 KQ461155 KPJ08165.1 KQ459232 KPJ02770.1 AGBW02009056 OWR51785.1 DS232161 GFDL01009848 JAV25197.1 GEZM01068259 JAV67065.1 CH477286 ADMH02000444 ETN66459.1 JXUM01067886 KQ562472 KXJ75822.1 AAAB01008980 EAA13916.4 CH916371 EDV92253.1 AXCM01000552 APCN01001664 CP012528 ALC49037.1 CH964239 EDW82534.1 CH933811 EDW06396.1 GBXI01009457 GBXI01007405 JAD04835.1 JAD06887.1 BT150439 AHV85501.1 AF017777 AE014298 BT003552 AAO39556.1 GDHF01026205 GDHF01023022 GDHF01020170 GDHF01011116 GDHF01010385 GDHF01001301 JAI26109.1 JAI29292.1 JAI32144.1 JAI41198.1 JAI41929.1 JAI51013.1 AXCN02001967 CH940655 EDW66406.2 CH379063 EDY72069.2 CM000366 EDX18534.1 CH479636 EDW35356.1 CH954180 EDV47617.2 CH902622 EDV34515.2 CH480823 EDW56378.1 CM000162 EDX03095.2 CCAG010004527 CCAG010004528 OUUW01000003 SPP77812.1 JXJN01001483 GAMC01012594 GAMC01012590 JAB93961.1 CVRI01000043 CRK96439.1 KB632308 ERL91963.1 KK107941 QOIP01000004 EZA47093.1 RLU23968.1 GAKP01006927 JAC52025.1 AJVK01015784 APGK01051610 KB741197 ENN72980.1 KQ981204 KYN44944.1 GDKW01001741 JAI54854.1 ACPB03015116 ENN72981.1 GDIQ01184955 JAK66770.1 GL732541 EFX81974.1 GBYB01008055 JAG77822.1 GDIQ01136766 JAL14960.1 GBGD01000893 JAC87996.1 KQ980761 KYN13602.1 GDIP01238823 JAI84578.1 GDIQ01042559 JAN52178.1 GBBI01002620 JAC16092.1 GDIQ01205261 JAK46464.1 LBMM01002135 KMQ95220.1 GL437912 EFN69903.1 GDIP01211684 JAJ11718.1 GL771866 EFZ09158.1 GL451091 EFN80045.1 GDIQ01101444 JAL50282.1 GDRN01066268 GDRN01066267 GDRN01066266 JAI64571.1 KQ976462 KYM84642.1 ADTU01003520 ADTU01003521 GDIP01163433 JAJ59969.1 GDIP01074452 JAM29263.1 AFYH01140933 AFYH01140934 AFYH01140935 AFYH01140936 GDIP01023074 JAM80641.1 GL888216 EGI64823.1 LRGB01002121 KZS09009.1 AERX01026919 AERX01026920 FR905222 CDQ77330.1 KQ761108 OAD58203.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000002320 UP000008820 UP000000673 UP000069940 UP000249989 UP000075901 UP000076407 UP000075900 UP000075882 UP000075885 UP000075881 UP000007062 UP000075920 UP000001070 UP000075883 UP000075902 UP000075880 UP000075840 UP000092553 UP000075903 UP000069272 UP000007798 UP000009192 UP000075884 UP000000803 UP000075886 UP000095301 UP000008792 UP000001819 UP000000304 UP000008744 UP000008711 UP000007801 UP000001292 UP000002282 UP000092444 UP000092443 UP000268350 UP000092445 UP000095300 UP000092460 UP000076408 UP000091820 UP000183832 UP000078200 UP000030742 UP000053097 UP000279307 UP000092462 UP000019118 UP000078541 UP000015103 UP000000305 UP000078492 UP000036403 UP000000311 UP000008237 UP000078540 UP000005205 UP000008672 UP000007755 UP000076858 UP000005207 UP000087266 UP000193380 UP000265160 UP000221080
UP000002320 UP000008820 UP000000673 UP000069940 UP000249989 UP000075901 UP000076407 UP000075900 UP000075882 UP000075885 UP000075881 UP000007062 UP000075920 UP000001070 UP000075883 UP000075902 UP000075880 UP000075840 UP000092553 UP000075903 UP000069272 UP000007798 UP000009192 UP000075884 UP000000803 UP000075886 UP000095301 UP000008792 UP000001819 UP000000304 UP000008744 UP000008711 UP000007801 UP000001292 UP000002282 UP000092444 UP000092443 UP000268350 UP000092445 UP000095300 UP000092460 UP000076408 UP000091820 UP000183832 UP000078200 UP000030742 UP000053097 UP000279307 UP000092462 UP000019118 UP000078541 UP000015103 UP000000305 UP000078492 UP000036403 UP000000311 UP000008237 UP000078540 UP000005205 UP000008672 UP000007755 UP000076858 UP000005207 UP000087266 UP000193380 UP000265160 UP000221080
Pfam
Interpro
IPR009000
Transl_B-barrel_sf
+ More
IPR006297 EF-4
IPR035647 EFG_III/V
IPR027417 P-loop_NTPase
IPR013842 LepA_CTD
IPR035654 LepA_IV
IPR000795 TF_GTP-bd_dom
IPR038363 LepA_C_sf
IPR000640 EFG_V-like
IPR005225 Small_GTP-bd_dom
IPR031157 G_TR_CS
IPR019164 TMEM147
IPR001320 Iontro_rcpt
IPR004161 EFTu-like_2
IPR006297 EF-4
IPR035647 EFG_III/V
IPR027417 P-loop_NTPase
IPR013842 LepA_CTD
IPR035654 LepA_IV
IPR000795 TF_GTP-bd_dom
IPR038363 LepA_C_sf
IPR000640 EFG_V-like
IPR005225 Small_GTP-bd_dom
IPR031157 G_TR_CS
IPR019164 TMEM147
IPR001320 Iontro_rcpt
IPR004161 EFTu-like_2
Gene 3D
CDD
ProteinModelPortal
H9JCM0
A0A2H1W0S4
A0A2A4JLQ2
A0A3S2P9K5
A0A194QT37
A0A194QCR0
+ More
A0A212FDE4 B0WXB8 A0A1Q3FCB3 A0A1Y1L517 Q0IFX5 W5JUN6 A0A182GJD8 A0A182T5N1 A0A182XGW6 A0A182RWS6 A0A182KZ19 A0A182PCG3 A0A182KGD9 Q7PMF6 A0A182W8T0 A0A1S4H2L6 B4JNG0 A0A182M5S3 A0A182TJP4 A0A182IZJ7 A0A182HRB1 A0A0M4F967 A0A182UQQ6 A0A182F0T2 B4NCH5 B4L5D7 A0A182NP40 A0A0A1X703 X4YDX6 Q9VRH6 Q86P01 A0A0K8VQR5 A0A182Q138 A0A1I8MJZ7 B4MAZ7 B5DKL9 B4R3R6 B4HD31 B3NYD8 B3MRW0 B4I6H2 B4PZ34 A0A1B0FCK3 A0A1A9YNV1 A0A3B0JCJ5 A0A1B0AEB4 A0A1I8PR26 A0A1B0APL7 W8BL04 A0A182Y6B8 A0A1A9X1G1 A0A1J1I7Z7 A0A1A9UL95 U4UHZ1 A0A026VTH6 A0A034WD04 A0A1B0DJF5 N6TUM5 A0A195FYN7 A0A0P4VQT4 T1HN81 N6TX65 A0A0N8BER3 E9GF27 A0A0C9RII1 A0A0P5QE36 A0A069DVY7 A0A151IYU8 A0A0P4XS62 A0A0P6GQK2 A0A023F4I2 A0A0P5J3B2 A0A0J7NRZ9 E2A9H9 A0A0P4ZJX9 E9JD47 E2BW13 A0A0P5RFK9 A0A0P4WS90 A0A151I492 A0A158NXT4 A0A0N8ABL3 A0A0P5XBV6 H3AGD0 A0A0P6AZP7 F4WLU1 A0A164RWH3 I3KFH1 A0A1S3RR63 A0A060XKC1 H3AGD1 A0A3P9BCX5 A0A310SDA6 A0A2D0RF47
A0A212FDE4 B0WXB8 A0A1Q3FCB3 A0A1Y1L517 Q0IFX5 W5JUN6 A0A182GJD8 A0A182T5N1 A0A182XGW6 A0A182RWS6 A0A182KZ19 A0A182PCG3 A0A182KGD9 Q7PMF6 A0A182W8T0 A0A1S4H2L6 B4JNG0 A0A182M5S3 A0A182TJP4 A0A182IZJ7 A0A182HRB1 A0A0M4F967 A0A182UQQ6 A0A182F0T2 B4NCH5 B4L5D7 A0A182NP40 A0A0A1X703 X4YDX6 Q9VRH6 Q86P01 A0A0K8VQR5 A0A182Q138 A0A1I8MJZ7 B4MAZ7 B5DKL9 B4R3R6 B4HD31 B3NYD8 B3MRW0 B4I6H2 B4PZ34 A0A1B0FCK3 A0A1A9YNV1 A0A3B0JCJ5 A0A1B0AEB4 A0A1I8PR26 A0A1B0APL7 W8BL04 A0A182Y6B8 A0A1A9X1G1 A0A1J1I7Z7 A0A1A9UL95 U4UHZ1 A0A026VTH6 A0A034WD04 A0A1B0DJF5 N6TUM5 A0A195FYN7 A0A0P4VQT4 T1HN81 N6TX65 A0A0N8BER3 E9GF27 A0A0C9RII1 A0A0P5QE36 A0A069DVY7 A0A151IYU8 A0A0P4XS62 A0A0P6GQK2 A0A023F4I2 A0A0P5J3B2 A0A0J7NRZ9 E2A9H9 A0A0P4ZJX9 E9JD47 E2BW13 A0A0P5RFK9 A0A0P4WS90 A0A151I492 A0A158NXT4 A0A0N8ABL3 A0A0P5XBV6 H3AGD0 A0A0P6AZP7 F4WLU1 A0A164RWH3 I3KFH1 A0A1S3RR63 A0A060XKC1 H3AGD1 A0A3P9BCX5 A0A310SDA6 A0A2D0RF47
PDB
2YWH
E-value=1.50925e-143,
Score=1308
Ontologies
GO
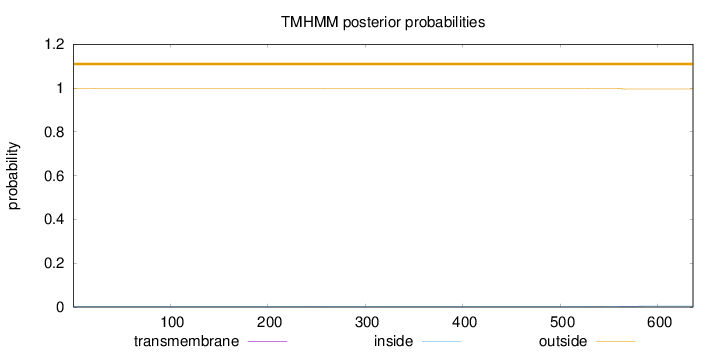
Topology
Subcellular location
Mitochondrion inner membrane
Length:
636
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.07931
Exp number, first 60 AAs:
0.00185
Total prob of N-in:
0.00155
outside
1 - 636
Population Genetic Test Statistics
Pi
228.115229
Theta
165.22659
Tajima's D
0.988699
CLR
0.261872
CSRT
0.653917304134793
Interpretation
Uncertain
