Pre Gene Modal
BGIBMGA007298
Annotation
putative_Wdr45l_protein_[Danaus_plexippus]
Full name
WD repeat domain phosphoinositide-interacting protein 3
Alternative Name
WD repeat-containing protein 45-like
WD repeat-containing protein 45B
WD repeat-containing protein 45B
Location in the cell
Extracellular Reliability : 2.029
Sequence
CDS
ATGAACTTGTCTGAAGAAAATCCATTTGGTAACGGGCTTTTGTACGTGGGCTTTAACCAAGATCAAGGTTGTTTCGCTTGTGGAACGGAAAATGGTTTCCGTGTCTTCAATAGTGACCCCGTACAAGAGAAAGAACGGCAGAACTTTGCTGTTGGCGGTCTTTCGTACGTGGAAATGTTATTCCGGTGTAATTATATGGCTTTGGTGGGAGGTGGTAAAACACCGGTCTATCCTCCGAACAAAGTTATTATTTGGGATGATCTAAAGAAAGATTCCGCCATATCGCTTGACTTCAACAGCCCTGTGAAAGCTGTGAAATTGCGGAGAGACCGTATTGTTGTTGTTCTAGAACAATTGATAAAAGTGTATACTTTTACGGCTCAGCCCCAATTGCTCCATGTGTTTGAAACTTGTCAAAACACAAAAGGGTTGTGCGTAGTATGTCCTAACAGTAATAATGCTCTTCTTGCCTATCCCAGCCGAAAAACTGGTCATGTGCAGTTAGTAGAGTTAAGTAGTCATGCAGGGCCAAGCACAGCCCCAGAAAGTCATCTAATAGCAGCACATGAGGCCCCTCTATGTTGTTTGGCACTTAATGTTGGAGGTACCAGACTAGCCACTGCTTCGACTAAAGGAACCCTAATAAGGGTTTTTGATACATCAACAGGACAGAAATTGGCTGAATTGCGAAGAGGAGCTCATCAGGCAACTATATACTGCATTAATTTTAACCACACAAGTACAAATCTTTGTGTGACCTCAGATCATGGAACTGTTCATGTATTCAGTTTAGAAGATGAGAAACTCAACAAACAGTCCACTTTGGCCACTGTTAACTTTTTGCCTAAATACTTCTCAAGCAATTGGAGTTTTTGCAAATTCACAATACCAAATGGTCCTCCCTGCATTTGTGCGTTTGGTGTTGACAAGAGTTCTATTATTGTGGTTTGTGCTGACGGCTCTTACTACAAATACAAATTCAATGAAAAGGGGGAATGCACCAGAGATGTTTATGATCAGTTTTTGGAGACTTCTGAAGATAGATAA
Protein
MNLSEENPFGNGLLYVGFNQDQGCFACGTENGFRVFNSDPVQEKERQNFAVGGLSYVEMLFRCNYMALVGGGKTPVYPPNKVIIWDDLKKDSAISLDFNSPVKAVKLRRDRIVVVLEQLIKVYTFTAQPQLLHVFETCQNTKGLCVVCPNSNNALLAYPSRKTGHVQLVELSSHAGPSTAPESHLIAAHEAPLCCLALNVGGTRLATASTKGTLIRVFDTSTGQKLAELRRGAHQATIYCINFNHTSTNLCVTSDHGTVHVFSLEDEKLNKQSTLATVNFLPKYFSSNWSFCKFTIPNGPPCICAFGVDKSSIIVVCADGSYYKYKFNEKGECTRDVYDQFLETSEDR
Summary
Description
Component of the autophagy machinery that controls the major intracellular degradation process by which cytoplasmic materials are packaged into autophagosomes and delivered to lysosomes for degradation. In the cellular response to starvation, may also function in the inhibition of the mTORC1 signaling pathway.
Similarity
Belongs to the WD repeat SVP1 family.
Keywords
Autophagy
Lysosome
Repeat
WD repeat
Complete proteome
Reference proteome
Feature
chain WD repeat domain phosphoinositide-interacting protein 3
Uniprot
H9JCQ1
A0A2A4JM05
A0A2H1W0L9
A0A3S2N9M7
A0A1E1WBQ8
A0A212FDI1
+ More
A0A194QS70 A0A194QH51 A0A0L7LLB0 A0A2J7QN31 K7IZI5 A0A1Y1LXH8 A0A067QMA3 A0A3L8E5Q4 E2BQR1 D6WYC7 E2AYD7 F4WD07 A0A195BVW5 A0A158NKN7 A0A1S3J2E5 A0A232FKR4 T1JCL6 A0A154PPM5 A0A1B6CXJ1 V9IH27 A0A088AEF1 A0A2A3E648 A0A0N0U3D8 E0VN38 A0A1L8EN29 Q68F45 V3ZIP6 A0A1S3DM49 A0A1B6IYL7 A0A0P4VMP5 T1I7K3 E9JBZ3 A0A1B6MPL6 A0A1B8Y0G6 Q640T2 A0A023F5H2 K1PZL8 A0A146MGD7 A0A0A9YY90 A0A1B6F4R1 A0A2C9JG06 A0A1A8IMF9 A0A1A8UV92 A0A1A8QUH7 A0A1A8LAX9 A0A1A8DZI0 A0A131Y5B6 A0A224XML6 A0A1Q3FDQ0 A0A1E1XF07 A0A1E1XUN7 A0A023FKV1 A0A3Q3N036 A0A0P7YDW4 A0A3Q3GUL9 K9IIX2 A0A3B3SGR7 A0A151NVT5 B0WR49 A0A2U9CKU7 A0A1A7WQQ0 A0A2L2Y9I4 A0A3B1IXZ5 A0A3B4EK78 A0A091IB64 A0A182GWD0 A0A3Q1EMQ4 A0A3B4FQI9 A0A3B5BHU6 A0A3Q2VXV9 A0A3Q4HFB3 A0A3Q1AJX3 A0A3Q0T7N3 I3KTX2 A0A3P8R4T9 A0A3P9B4D5 A0A3P8TKS9 Q5ZL16 G3PR41 U3DI25 A0A3Q2ZL05 W5UCN0 A0A3B4YMV3 A0A3B3X8X9 A0A3B5PS17 A0A3Q1JUG6 A0A087YMH1 A0A3B4UHP4 A0A3B3VXE6 A8KC31 J3S9X4 A0A0K8TNI9 A0A286X7P3 A0A091DUG4
A0A194QS70 A0A194QH51 A0A0L7LLB0 A0A2J7QN31 K7IZI5 A0A1Y1LXH8 A0A067QMA3 A0A3L8E5Q4 E2BQR1 D6WYC7 E2AYD7 F4WD07 A0A195BVW5 A0A158NKN7 A0A1S3J2E5 A0A232FKR4 T1JCL6 A0A154PPM5 A0A1B6CXJ1 V9IH27 A0A088AEF1 A0A2A3E648 A0A0N0U3D8 E0VN38 A0A1L8EN29 Q68F45 V3ZIP6 A0A1S3DM49 A0A1B6IYL7 A0A0P4VMP5 T1I7K3 E9JBZ3 A0A1B6MPL6 A0A1B8Y0G6 Q640T2 A0A023F5H2 K1PZL8 A0A146MGD7 A0A0A9YY90 A0A1B6F4R1 A0A2C9JG06 A0A1A8IMF9 A0A1A8UV92 A0A1A8QUH7 A0A1A8LAX9 A0A1A8DZI0 A0A131Y5B6 A0A224XML6 A0A1Q3FDQ0 A0A1E1XF07 A0A1E1XUN7 A0A023FKV1 A0A3Q3N036 A0A0P7YDW4 A0A3Q3GUL9 K9IIX2 A0A3B3SGR7 A0A151NVT5 B0WR49 A0A2U9CKU7 A0A1A7WQQ0 A0A2L2Y9I4 A0A3B1IXZ5 A0A3B4EK78 A0A091IB64 A0A182GWD0 A0A3Q1EMQ4 A0A3B4FQI9 A0A3B5BHU6 A0A3Q2VXV9 A0A3Q4HFB3 A0A3Q1AJX3 A0A3Q0T7N3 I3KTX2 A0A3P8R4T9 A0A3P9B4D5 A0A3P8TKS9 Q5ZL16 G3PR41 U3DI25 A0A3Q2ZL05 W5UCN0 A0A3B4YMV3 A0A3B3X8X9 A0A3B5PS17 A0A3Q1JUG6 A0A087YMH1 A0A3B4UHP4 A0A3B3VXE6 A8KC31 J3S9X4 A0A0K8TNI9 A0A286X7P3 A0A091DUG4
Pubmed
19121390
22118469
26354079
26227816
20075255
28004739
+ More
24845553 30249741 20798317 18362917 19820115 21719571 21347285 28648823 20566863 27762356 23254933 27129103 21282665 20431018 25474469 22992520 26823975 25401762 15562597 29652888 28503490 29209593 29240929 22293439 26561354 25329095 26483478 25186727 15642098 25243066 23127152 23542700 12477932 23594743 23025625 26369729 21993624
24845553 30249741 20798317 18362917 19820115 21719571 21347285 28648823 20566863 27762356 23254933 27129103 21282665 20431018 25474469 22992520 26823975 25401762 15562597 29652888 28503490 29209593 29240929 22293439 26561354 25329095 26483478 25186727 15642098 25243066 23127152 23542700 12477932 23594743 23025625 26369729 21993624
EMBL
BABH01034693
NWSH01001074
PCG72748.1
ODYU01005610
SOQ46628.1
RSAL01000168
+ More
RVE45235.1 GDQN01006679 JAT84375.1 AGBW02009056 OWR51784.1 KQ461155 KPJ08164.1 KQ459232 KPJ02771.1 JTDY01000686 KOB76222.1 NEVH01013201 PNF29994.1 GEZM01048841 JAV76356.1 KK853177 KDR10271.1 QOIP01000001 RLU27178.1 GL449798 EFN81980.1 KQ971356 EFA09113.1 GL443887 EFN61554.1 GL888077 EGI67914.1 KQ976403 KYM92113.1 ADTU01002112 NNAY01000090 OXU31089.1 JH432065 KQ435012 KZC13819.1 GEDC01019124 GEDC01001200 JAS18174.1 JAS36098.1 JR047272 AEY60370.1 KZ288355 PBC27217.1 KQ435908 KOX68971.1 DS235332 EEB14804.1 CM004483 OCT60763.1 BC080000 KB203566 ESO84102.1 GECU01015681 JAS92025.1 GDKW01000761 JAI55834.1 ACPB03000177 ACPB03000178 GL771642 EFZ09638.1 GEBQ01002077 JAT37900.1 KV460620 OCA16386.1 CR762340 BC082507 GBBI01002314 JAC16398.1 JH817657 EKC29662.1 GDHC01000899 JAQ17730.1 GBHO01009072 GBHO01009071 GBHO01009070 GBHO01009069 GBHO01009068 GBHO01009067 GBRD01012034 JAG34532.1 JAG34533.1 JAG34534.1 JAG34535.1 JAG34536.1 JAG34537.1 JAG53790.1 GECZ01024591 JAS45178.1 HAED01012056 SBQ98450.1 HAEJ01010521 SBS50978.1 HAEI01006232 SBR96694.1 HAEF01004134 HAEG01013132 SBR41516.1 HADZ01007717 HAEA01010852 SBQ39332.1 GEFM01002675 GEGO01001600 JAP73121.1 JAR93804.1 GFTR01005358 JAW11068.1 GFDL01009437 JAV25608.1 GFAC01001341 JAT97847.1 GFAA01000675 JAU02760.1 GBBK01002847 JAC21635.1 JARO02006984 KPP64515.1 GABZ01006857 JAA46668.1 AKHW03001857 KYO40843.1 DS232051 EDS33155.1 CP026260 AWP17224.1 HADW01006645 HADX01002953 SBP08045.1 IAAA01009632 IAAA01009633 IAAA01009634 LAA04010.1 KL218461 KFP05447.1 JXUM01004319 JXUM01004320 KQ560172 KXJ84013.1 AERX01002006 AERX01002007 AJ719918 GAMT01008559 GAMS01010125 GAMR01001085 GAMQ01006835 GAMP01005637 JAB03302.1 JAB13011.1 JAB32847.1 JAB35016.1 JAB47118.1 JT406127 AHH37467.1 AYCK01000860 CU611036 BC154337 AAI54338.1 JU176545 GBEX01003994 AFJ52068.1 JAI10566.1 GDAI01001900 JAI15703.1 AAKN02047362 KN121670 KFO35744.1
RVE45235.1 GDQN01006679 JAT84375.1 AGBW02009056 OWR51784.1 KQ461155 KPJ08164.1 KQ459232 KPJ02771.1 JTDY01000686 KOB76222.1 NEVH01013201 PNF29994.1 GEZM01048841 JAV76356.1 KK853177 KDR10271.1 QOIP01000001 RLU27178.1 GL449798 EFN81980.1 KQ971356 EFA09113.1 GL443887 EFN61554.1 GL888077 EGI67914.1 KQ976403 KYM92113.1 ADTU01002112 NNAY01000090 OXU31089.1 JH432065 KQ435012 KZC13819.1 GEDC01019124 GEDC01001200 JAS18174.1 JAS36098.1 JR047272 AEY60370.1 KZ288355 PBC27217.1 KQ435908 KOX68971.1 DS235332 EEB14804.1 CM004483 OCT60763.1 BC080000 KB203566 ESO84102.1 GECU01015681 JAS92025.1 GDKW01000761 JAI55834.1 ACPB03000177 ACPB03000178 GL771642 EFZ09638.1 GEBQ01002077 JAT37900.1 KV460620 OCA16386.1 CR762340 BC082507 GBBI01002314 JAC16398.1 JH817657 EKC29662.1 GDHC01000899 JAQ17730.1 GBHO01009072 GBHO01009071 GBHO01009070 GBHO01009069 GBHO01009068 GBHO01009067 GBRD01012034 JAG34532.1 JAG34533.1 JAG34534.1 JAG34535.1 JAG34536.1 JAG34537.1 JAG53790.1 GECZ01024591 JAS45178.1 HAED01012056 SBQ98450.1 HAEJ01010521 SBS50978.1 HAEI01006232 SBR96694.1 HAEF01004134 HAEG01013132 SBR41516.1 HADZ01007717 HAEA01010852 SBQ39332.1 GEFM01002675 GEGO01001600 JAP73121.1 JAR93804.1 GFTR01005358 JAW11068.1 GFDL01009437 JAV25608.1 GFAC01001341 JAT97847.1 GFAA01000675 JAU02760.1 GBBK01002847 JAC21635.1 JARO02006984 KPP64515.1 GABZ01006857 JAA46668.1 AKHW03001857 KYO40843.1 DS232051 EDS33155.1 CP026260 AWP17224.1 HADW01006645 HADX01002953 SBP08045.1 IAAA01009632 IAAA01009633 IAAA01009634 LAA04010.1 KL218461 KFP05447.1 JXUM01004319 JXUM01004320 KQ560172 KXJ84013.1 AERX01002006 AERX01002007 AJ719918 GAMT01008559 GAMS01010125 GAMR01001085 GAMQ01006835 GAMP01005637 JAB03302.1 JAB13011.1 JAB32847.1 JAB35016.1 JAB47118.1 JT406127 AHH37467.1 AYCK01000860 CU611036 BC154337 AAI54338.1 JU176545 GBEX01003994 AFJ52068.1 JAI10566.1 GDAI01001900 JAI15703.1 AAKN02047362 KN121670 KFO35744.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000053240
UP000053268
+ More
UP000037510 UP000235965 UP000002358 UP000027135 UP000279307 UP000008237 UP000007266 UP000000311 UP000007755 UP000078540 UP000005205 UP000085678 UP000215335 UP000076502 UP000005203 UP000242457 UP000053105 UP000009046 UP000186698 UP000030746 UP000079169 UP000015103 UP000008143 UP000005408 UP000076420 UP000261640 UP000034805 UP000192224 UP000261660 UP000261540 UP000050525 UP000002320 UP000246464 UP000018467 UP000261440 UP000054308 UP000069940 UP000249989 UP000257200 UP000261460 UP000261400 UP000264840 UP000261580 UP000257160 UP000261340 UP000005207 UP000265100 UP000265160 UP000265080 UP000000539 UP000007635 UP000008225 UP000264800 UP000221080 UP000261360 UP000261480 UP000002852 UP000265040 UP000028760 UP000261420 UP000261500 UP000000437 UP000005447 UP000028990
UP000037510 UP000235965 UP000002358 UP000027135 UP000279307 UP000008237 UP000007266 UP000000311 UP000007755 UP000078540 UP000005205 UP000085678 UP000215335 UP000076502 UP000005203 UP000242457 UP000053105 UP000009046 UP000186698 UP000030746 UP000079169 UP000015103 UP000008143 UP000005408 UP000076420 UP000261640 UP000034805 UP000192224 UP000261660 UP000261540 UP000050525 UP000002320 UP000246464 UP000018467 UP000261440 UP000054308 UP000069940 UP000249989 UP000257200 UP000261460 UP000261400 UP000264840 UP000261580 UP000257160 UP000261340 UP000005207 UP000265100 UP000265160 UP000265080 UP000000539 UP000007635 UP000008225 UP000264800 UP000221080 UP000261360 UP000261480 UP000002852 UP000265040 UP000028760 UP000261420 UP000261500 UP000000437 UP000005447 UP000028990
Pfam
PF00400 WD40
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
H9JCQ1
A0A2A4JM05
A0A2H1W0L9
A0A3S2N9M7
A0A1E1WBQ8
A0A212FDI1
+ More
A0A194QS70 A0A194QH51 A0A0L7LLB0 A0A2J7QN31 K7IZI5 A0A1Y1LXH8 A0A067QMA3 A0A3L8E5Q4 E2BQR1 D6WYC7 E2AYD7 F4WD07 A0A195BVW5 A0A158NKN7 A0A1S3J2E5 A0A232FKR4 T1JCL6 A0A154PPM5 A0A1B6CXJ1 V9IH27 A0A088AEF1 A0A2A3E648 A0A0N0U3D8 E0VN38 A0A1L8EN29 Q68F45 V3ZIP6 A0A1S3DM49 A0A1B6IYL7 A0A0P4VMP5 T1I7K3 E9JBZ3 A0A1B6MPL6 A0A1B8Y0G6 Q640T2 A0A023F5H2 K1PZL8 A0A146MGD7 A0A0A9YY90 A0A1B6F4R1 A0A2C9JG06 A0A1A8IMF9 A0A1A8UV92 A0A1A8QUH7 A0A1A8LAX9 A0A1A8DZI0 A0A131Y5B6 A0A224XML6 A0A1Q3FDQ0 A0A1E1XF07 A0A1E1XUN7 A0A023FKV1 A0A3Q3N036 A0A0P7YDW4 A0A3Q3GUL9 K9IIX2 A0A3B3SGR7 A0A151NVT5 B0WR49 A0A2U9CKU7 A0A1A7WQQ0 A0A2L2Y9I4 A0A3B1IXZ5 A0A3B4EK78 A0A091IB64 A0A182GWD0 A0A3Q1EMQ4 A0A3B4FQI9 A0A3B5BHU6 A0A3Q2VXV9 A0A3Q4HFB3 A0A3Q1AJX3 A0A3Q0T7N3 I3KTX2 A0A3P8R4T9 A0A3P9B4D5 A0A3P8TKS9 Q5ZL16 G3PR41 U3DI25 A0A3Q2ZL05 W5UCN0 A0A3B4YMV3 A0A3B3X8X9 A0A3B5PS17 A0A3Q1JUG6 A0A087YMH1 A0A3B4UHP4 A0A3B3VXE6 A8KC31 J3S9X4 A0A0K8TNI9 A0A286X7P3 A0A091DUG4
A0A194QS70 A0A194QH51 A0A0L7LLB0 A0A2J7QN31 K7IZI5 A0A1Y1LXH8 A0A067QMA3 A0A3L8E5Q4 E2BQR1 D6WYC7 E2AYD7 F4WD07 A0A195BVW5 A0A158NKN7 A0A1S3J2E5 A0A232FKR4 T1JCL6 A0A154PPM5 A0A1B6CXJ1 V9IH27 A0A088AEF1 A0A2A3E648 A0A0N0U3D8 E0VN38 A0A1L8EN29 Q68F45 V3ZIP6 A0A1S3DM49 A0A1B6IYL7 A0A0P4VMP5 T1I7K3 E9JBZ3 A0A1B6MPL6 A0A1B8Y0G6 Q640T2 A0A023F5H2 K1PZL8 A0A146MGD7 A0A0A9YY90 A0A1B6F4R1 A0A2C9JG06 A0A1A8IMF9 A0A1A8UV92 A0A1A8QUH7 A0A1A8LAX9 A0A1A8DZI0 A0A131Y5B6 A0A224XML6 A0A1Q3FDQ0 A0A1E1XF07 A0A1E1XUN7 A0A023FKV1 A0A3Q3N036 A0A0P7YDW4 A0A3Q3GUL9 K9IIX2 A0A3B3SGR7 A0A151NVT5 B0WR49 A0A2U9CKU7 A0A1A7WQQ0 A0A2L2Y9I4 A0A3B1IXZ5 A0A3B4EK78 A0A091IB64 A0A182GWD0 A0A3Q1EMQ4 A0A3B4FQI9 A0A3B5BHU6 A0A3Q2VXV9 A0A3Q4HFB3 A0A3Q1AJX3 A0A3Q0T7N3 I3KTX2 A0A3P8R4T9 A0A3P9B4D5 A0A3P8TKS9 Q5ZL16 G3PR41 U3DI25 A0A3Q2ZL05 W5UCN0 A0A3B4YMV3 A0A3B3X8X9 A0A3B5PS17 A0A3Q1JUG6 A0A087YMH1 A0A3B4UHP4 A0A3B3VXE6 A8KC31 J3S9X4 A0A0K8TNI9 A0A286X7P3 A0A091DUG4
PDB
6IYY
E-value=4.15327e-124,
Score=1138
Ontologies
GO
Topology
Subcellular location
Preautophagosomal structure
Lysosome
Lysosome
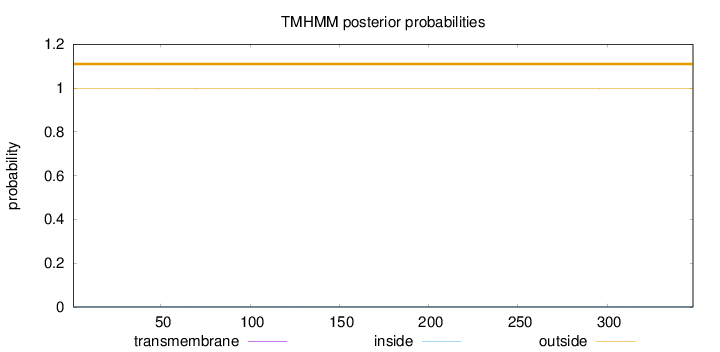
Length:
348
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02349
Exp number, first 60 AAs:
0.00491
Total prob of N-in:
0.00355
outside
1 - 348
Population Genetic Test Statistics
Pi
231.970092
Theta
215.186583
Tajima's D
0.261006
CLR
0
CSRT
0.44272786360682
Interpretation
Uncertain
