Gene
KWMTBOMO10290
Annotation
hypothetical_protein_RR46_09970_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 1.022 Mitochondrial Reliability : 1.326 Nuclear Reliability : 1.32
Sequence
CDS
ATGAAGAAATACGGATCCCGCAATGCTTTATATGATAAGCTGCACAAGGGTGGTGTATTAGTTTGCATCGGATTGACATTGTATGGGACTGTTCTACTCACTGATCACTTTTACAAGTACTTCAAGTATGTGCGGCCACAGATACAAGCATCCAAAGCAGCTGCTGAACAAGAGCTTTTGTCAGAAGGTTCTTCAGAGAAAATTATGTAA
Protein
MKKYGSRNALYDKLHKGGVLVCIGLTLYGTVLLTDHFYKYFKYVRPQIQASKAAAEQELLSEGSSEKIM
Summary
Uniprot
A0A3S2TGC8
A0A2H1W287
A0A194QBD1
A0A194QX66
A0A0L7K3K2
A0A212FDL2
+ More
S4PWS1 A0A1W4WH43 A0A0M4EIR4 A0A2J7QGA2 B4JLC1 A0A3B0JTC1 E2C5C5 B4I9R3 B3MYS0 A0A0R1EA09 B4M6M8 Q29JL8 B4H379 B4Q195 B3P8X9 A0A2P8XPN9 A0A067QL80 A0A195ERJ3 A0A1W4VEI6 B4NQ12 A0A1B0DK92 Q16YG1 B4L200 A0A023EBF9 A0A023EDG8 A0A182VXA6 A0A1L8DZV1 Q9W513 A0A026W877 Q4QPS4 A0A182M8L1 R4V366 A0A182HT28 A0A0J7L5W4 A0A0L0C371 A0A182V955 A0A182K8T8 A0A182XI46 A0A182LF72 Q7PGH2 A0A1L8EBR5 U5EQH1 A0A1B0CV88 A0A195D862 E1ZWI8 A0A1I8M2U9 A0A182XWZ6 A0A151X3I5 A0A084VU60 A0A182QD97 A0A1I8QE26 A0A2M4CY79 W5JFH1 A0A2M4AWX4 A0A2M4C5A1 A0A195CZR5 A0A182NFH4 A0A2M3ZKP9 A0A1A9WWY1 T1DNW1 A0A195BRI6 A0A1J1HGB4 A0A2A3EQI6 A0A0K8TPD8 A0A226EEE7 A0A0L7RBC1 A0A1Q3FIC4 A0A1B0AXZ2 A0A1Q3FIM6 A0A088A7Z3 B0X6H0 A0A224XXT3 A0A1B0G816 A0A3S1CEA5 A0A182P742 A0A154PMC6 A0A183J6Z3 A0A085NQL9 E9FVV1 A0A336MZ32
S4PWS1 A0A1W4WH43 A0A0M4EIR4 A0A2J7QGA2 B4JLC1 A0A3B0JTC1 E2C5C5 B4I9R3 B3MYS0 A0A0R1EA09 B4M6M8 Q29JL8 B4H379 B4Q195 B3P8X9 A0A2P8XPN9 A0A067QL80 A0A195ERJ3 A0A1W4VEI6 B4NQ12 A0A1B0DK92 Q16YG1 B4L200 A0A023EBF9 A0A023EDG8 A0A182VXA6 A0A1L8DZV1 Q9W513 A0A026W877 Q4QPS4 A0A182M8L1 R4V366 A0A182HT28 A0A0J7L5W4 A0A0L0C371 A0A182V955 A0A182K8T8 A0A182XI46 A0A182LF72 Q7PGH2 A0A1L8EBR5 U5EQH1 A0A1B0CV88 A0A195D862 E1ZWI8 A0A1I8M2U9 A0A182XWZ6 A0A151X3I5 A0A084VU60 A0A182QD97 A0A1I8QE26 A0A2M4CY79 W5JFH1 A0A2M4AWX4 A0A2M4C5A1 A0A195CZR5 A0A182NFH4 A0A2M3ZKP9 A0A1A9WWY1 T1DNW1 A0A195BRI6 A0A1J1HGB4 A0A2A3EQI6 A0A0K8TPD8 A0A226EEE7 A0A0L7RBC1 A0A1Q3FIC4 A0A1B0AXZ2 A0A1Q3FIM6 A0A088A7Z3 B0X6H0 A0A224XXT3 A0A1B0G816 A0A3S1CEA5 A0A182P742 A0A154PMC6 A0A183J6Z3 A0A085NQL9 E9FVV1 A0A336MZ32
Pubmed
26354079
26227816
22118469
23622113
17994087
20798317
+ More
17550304 15632085 29403074 24845553 17510324 24945155 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 26108605 20966253 12364791 14747013 17210077 25315136 25244985 24438588 20920257 23761445 26369729 24929829 21292972
17550304 15632085 29403074 24845553 17510324 24945155 26483478 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24508170 26108605 20966253 12364791 14747013 17210077 25315136 25244985 24438588 20920257 23761445 26369729 24929829 21292972
EMBL
RSAL01000168
RVE45241.1
ODYU01005610
SOQ46634.1
KQ459232
KPJ02767.1
+ More
KQ461155 KPJ08166.1 JTDY01012630 KOB52244.1 AGBW02009056 OWR51788.1 GAIX01006298 JAA86262.1 CP012528 ALC48854.1 NEVH01014835 PNF27602.1 CH916370 EDW00374.1 OUUW01000003 SPP78710.1 GL452770 EFN76858.1 CH480825 EDW43944.1 CH902632 EDV32764.1 CM000162 KRK06139.1 CH940653 EDW62445.1 CH379063 EAL32283.1 CH479205 EDW30772.1 EDX01402.1 CH954183 EDV45584.1 PYGN01001573 PSN33966.1 KK853191 KDR10027.1 KQ981993 KYN30875.1 CH964291 EDW86237.1 AJVK01066748 CH477516 EAT39695.1 CH933810 EDW07721.1 JXUM01027213 GAPW01006805 GAPW01006804 KQ560783 JAC06793.1 JAC06794.1 KXJ80905.1 GAPW01006673 JAC06925.1 GFDF01002056 JAV12028.1 AE014298 KX531393 AAF45745.1 AHN59278.1 ANY27203.1 KK107405 EZA51224.1 BT023692 AAY85092.2 AXCM01012581 KC632414 AGM32228.1 APCN01000403 LBMM01000590 KMQ98006.1 JRES01000955 KNC26793.1 AAAB01008859 EAA44933.4 EGK96888.1 GFDG01002609 JAV16190.1 GANO01004252 JAB55619.1 AJWK01030266 KQ981153 KYN09063.1 GL434853 EFN74425.1 KQ982562 KYQ54951.1 ATLV01016645 KE525099 KFB41504.1 AXCN02000213 GGFL01005983 MBW70161.1 ADMH02001552 ETN62088.1 GGFK01011972 MBW45293.1 GGFJ01011361 MBW60502.1 KQ977041 KYN06153.1 GGFM01008321 MBW29072.1 GAMD01002515 JAA99075.1 KQ976424 KYM88564.1 CVRI01000002 CRK86915.1 KZ288203 PBC33399.1 GDAI01001843 JAI15760.1 LNIX01000004 OXA55437.1 KQ414617 KOC68164.1 GFDL01007714 JAV27331.1 JXJN01005462 GFDL01007713 JAV27332.1 DS232413 EDS41453.1 GFTR01000292 JAW16134.1 CCAG010000155 RQTK01000035 RUS90331.1 KQ434977 KZC12887.1 UZAM01016086 VDP41729.1 KL363639 KL367480 KN538437 KFD45215.1 KFD71765.1 KHJ40488.1 GL732525 EFX89039.1 UFQS01000611 UFQS01004199 UFQT01000611 UFQT01004199 SSX05380.1 SSX35566.1
KQ461155 KPJ08166.1 JTDY01012630 KOB52244.1 AGBW02009056 OWR51788.1 GAIX01006298 JAA86262.1 CP012528 ALC48854.1 NEVH01014835 PNF27602.1 CH916370 EDW00374.1 OUUW01000003 SPP78710.1 GL452770 EFN76858.1 CH480825 EDW43944.1 CH902632 EDV32764.1 CM000162 KRK06139.1 CH940653 EDW62445.1 CH379063 EAL32283.1 CH479205 EDW30772.1 EDX01402.1 CH954183 EDV45584.1 PYGN01001573 PSN33966.1 KK853191 KDR10027.1 KQ981993 KYN30875.1 CH964291 EDW86237.1 AJVK01066748 CH477516 EAT39695.1 CH933810 EDW07721.1 JXUM01027213 GAPW01006805 GAPW01006804 KQ560783 JAC06793.1 JAC06794.1 KXJ80905.1 GAPW01006673 JAC06925.1 GFDF01002056 JAV12028.1 AE014298 KX531393 AAF45745.1 AHN59278.1 ANY27203.1 KK107405 EZA51224.1 BT023692 AAY85092.2 AXCM01012581 KC632414 AGM32228.1 APCN01000403 LBMM01000590 KMQ98006.1 JRES01000955 KNC26793.1 AAAB01008859 EAA44933.4 EGK96888.1 GFDG01002609 JAV16190.1 GANO01004252 JAB55619.1 AJWK01030266 KQ981153 KYN09063.1 GL434853 EFN74425.1 KQ982562 KYQ54951.1 ATLV01016645 KE525099 KFB41504.1 AXCN02000213 GGFL01005983 MBW70161.1 ADMH02001552 ETN62088.1 GGFK01011972 MBW45293.1 GGFJ01011361 MBW60502.1 KQ977041 KYN06153.1 GGFM01008321 MBW29072.1 GAMD01002515 JAA99075.1 KQ976424 KYM88564.1 CVRI01000002 CRK86915.1 KZ288203 PBC33399.1 GDAI01001843 JAI15760.1 LNIX01000004 OXA55437.1 KQ414617 KOC68164.1 GFDL01007714 JAV27331.1 JXJN01005462 GFDL01007713 JAV27332.1 DS232413 EDS41453.1 GFTR01000292 JAW16134.1 CCAG010000155 RQTK01000035 RUS90331.1 KQ434977 KZC12887.1 UZAM01016086 VDP41729.1 KL363639 KL367480 KN538437 KFD45215.1 KFD71765.1 KHJ40488.1 GL732525 EFX89039.1 UFQS01000611 UFQS01004199 UFQT01000611 UFQT01004199 SSX05380.1 SSX35566.1
Proteomes
UP000283053
UP000053268
UP000053240
UP000037510
UP000007151
UP000192223
+ More
UP000092553 UP000235965 UP000001070 UP000268350 UP000008237 UP000001292 UP000007801 UP000002282 UP000008792 UP000001819 UP000008744 UP000008711 UP000245037 UP000027135 UP000078541 UP000192221 UP000007798 UP000092462 UP000008820 UP000009192 UP000069940 UP000249989 UP000075920 UP000000803 UP000053097 UP000075883 UP000075840 UP000036403 UP000037069 UP000075903 UP000075881 UP000076407 UP000075882 UP000007062 UP000092461 UP000078492 UP000000311 UP000095301 UP000076408 UP000075809 UP000030765 UP000075886 UP000095300 UP000000673 UP000078542 UP000075884 UP000091820 UP000078540 UP000183832 UP000242457 UP000198287 UP000053825 UP000092460 UP000005203 UP000002320 UP000092444 UP000271974 UP000075885 UP000076502 UP000050793 UP000000305
UP000092553 UP000235965 UP000001070 UP000268350 UP000008237 UP000001292 UP000007801 UP000002282 UP000008792 UP000001819 UP000008744 UP000008711 UP000245037 UP000027135 UP000078541 UP000192221 UP000007798 UP000092462 UP000008820 UP000009192 UP000069940 UP000249989 UP000075920 UP000000803 UP000053097 UP000075883 UP000075840 UP000036403 UP000037069 UP000075903 UP000075881 UP000076407 UP000075882 UP000007062 UP000092461 UP000078492 UP000000311 UP000095301 UP000076408 UP000075809 UP000030765 UP000075886 UP000095300 UP000000673 UP000078542 UP000075884 UP000091820 UP000078540 UP000183832 UP000242457 UP000198287 UP000053825 UP000092460 UP000005203 UP000002320 UP000092444 UP000271974 UP000075885 UP000076502 UP000050793 UP000000305
Interpro
SUPFAM
SSF54593
SSF54593
Gene 3D
CDD
ProteinModelPortal
A0A3S2TGC8
A0A2H1W287
A0A194QBD1
A0A194QX66
A0A0L7K3K2
A0A212FDL2
+ More
S4PWS1 A0A1W4WH43 A0A0M4EIR4 A0A2J7QGA2 B4JLC1 A0A3B0JTC1 E2C5C5 B4I9R3 B3MYS0 A0A0R1EA09 B4M6M8 Q29JL8 B4H379 B4Q195 B3P8X9 A0A2P8XPN9 A0A067QL80 A0A195ERJ3 A0A1W4VEI6 B4NQ12 A0A1B0DK92 Q16YG1 B4L200 A0A023EBF9 A0A023EDG8 A0A182VXA6 A0A1L8DZV1 Q9W513 A0A026W877 Q4QPS4 A0A182M8L1 R4V366 A0A182HT28 A0A0J7L5W4 A0A0L0C371 A0A182V955 A0A182K8T8 A0A182XI46 A0A182LF72 Q7PGH2 A0A1L8EBR5 U5EQH1 A0A1B0CV88 A0A195D862 E1ZWI8 A0A1I8M2U9 A0A182XWZ6 A0A151X3I5 A0A084VU60 A0A182QD97 A0A1I8QE26 A0A2M4CY79 W5JFH1 A0A2M4AWX4 A0A2M4C5A1 A0A195CZR5 A0A182NFH4 A0A2M3ZKP9 A0A1A9WWY1 T1DNW1 A0A195BRI6 A0A1J1HGB4 A0A2A3EQI6 A0A0K8TPD8 A0A226EEE7 A0A0L7RBC1 A0A1Q3FIC4 A0A1B0AXZ2 A0A1Q3FIM6 A0A088A7Z3 B0X6H0 A0A224XXT3 A0A1B0G816 A0A3S1CEA5 A0A182P742 A0A154PMC6 A0A183J6Z3 A0A085NQL9 E9FVV1 A0A336MZ32
S4PWS1 A0A1W4WH43 A0A0M4EIR4 A0A2J7QGA2 B4JLC1 A0A3B0JTC1 E2C5C5 B4I9R3 B3MYS0 A0A0R1EA09 B4M6M8 Q29JL8 B4H379 B4Q195 B3P8X9 A0A2P8XPN9 A0A067QL80 A0A195ERJ3 A0A1W4VEI6 B4NQ12 A0A1B0DK92 Q16YG1 B4L200 A0A023EBF9 A0A023EDG8 A0A182VXA6 A0A1L8DZV1 Q9W513 A0A026W877 Q4QPS4 A0A182M8L1 R4V366 A0A182HT28 A0A0J7L5W4 A0A0L0C371 A0A182V955 A0A182K8T8 A0A182XI46 A0A182LF72 Q7PGH2 A0A1L8EBR5 U5EQH1 A0A1B0CV88 A0A195D862 E1ZWI8 A0A1I8M2U9 A0A182XWZ6 A0A151X3I5 A0A084VU60 A0A182QD97 A0A1I8QE26 A0A2M4CY79 W5JFH1 A0A2M4AWX4 A0A2M4C5A1 A0A195CZR5 A0A182NFH4 A0A2M3ZKP9 A0A1A9WWY1 T1DNW1 A0A195BRI6 A0A1J1HGB4 A0A2A3EQI6 A0A0K8TPD8 A0A226EEE7 A0A0L7RBC1 A0A1Q3FIC4 A0A1B0AXZ2 A0A1Q3FIM6 A0A088A7Z3 B0X6H0 A0A224XXT3 A0A1B0G816 A0A3S1CEA5 A0A182P742 A0A154PMC6 A0A183J6Z3 A0A085NQL9 E9FVV1 A0A336MZ32
Ontologies
PANTHER
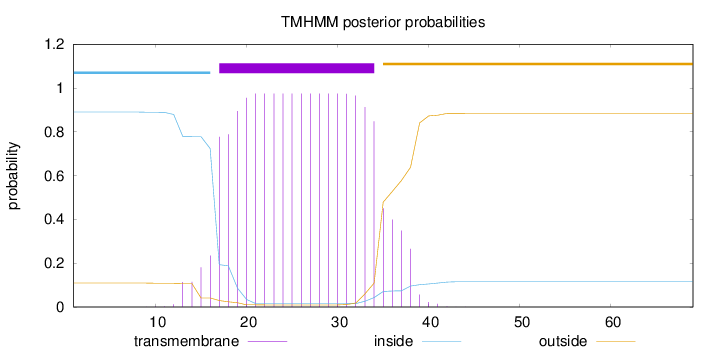
Topology
Length:
69
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.08709
Exp number, first 60 AAs:
19.08709
Total prob of N-in:
0.89053
POSSIBLE N-term signal
sequence
inside
1 - 16
TMhelix
17 - 34
outside
35 - 69
Population Genetic Test Statistics
Pi
17.673958
Theta
13.666746
Tajima's D
0.933754
CLR
0.083617
CSRT
0.649417529123544
Interpretation
Uncertain
