Gene
KWMTBOMO10289
Pre Gene Modal
BGIBMGA007296
Annotation
transposase_[Chilo_suppressalis]
Location in the cell
Nuclear Reliability : 2.939
Sequence
CDS
ATGTCTAAGCGGTTGGCAGAGCAAGATATTGAAAATATTTTATCAGTTCTCGAAAATGGAGACATTTCTGAAGATGAAGAGTCTATAGGAGACGAAAATGACATAGATTACTACTCGAATGTACAAGATCTGATACAGGATTTAGAAGATGCAGAAGAAGATCAGTATAATGCAAATCCTGATGTACCTATTGAAGAAGACCATCTACCGGATGAGTTTTCAGTTCCAGGGCCTTCACTTAGTCGCCCACTATCAGGTGTCATTGGTGCAGCAGCCTTGAGGCAACGAAGTAGGCAACTTATATGGAAAAAAGGCAATTTGGAATTTAGTGAAGACCGCATAAGTTTTACAGGTAATATAGACCTTTCCCCAGAATTTGCCCAACTAGAAACACCATTTCAGTATTTTACGCTATTTTTAAACGATGATATATTAGTAAAAATCGTGAATGAGACCAATTTGTATATCACGCAAAAAGGCATCTCCAACTGCCCTCCTGTAAATGTCAAGGAAATTCGTCAGTTTCTTGGTATTATTATATTTATGTCAGTATATCACTTCCCAAATGTAAGATCATATTGGGGTAAGTATGGCTTCAAACATATAATGGAAACGATGACGCTAAATAGATTTGGAAAACTACGATCCGTCATTCACTTCAATGACAACTCATTACACAAGCCCGTTGGTCACCCAAACCACGACCGTCTGCATAAAATAAGACCTGTAGAGGCACATTTGAATAAATTATTTTTGAGTGTGGCACCATTTGAGCAGAGACTGTCCTTAGATGAACAGATGTGCTCTACGAAAGTAGCGCATTTCATGAAACAATACCTCCCCAACAAGCCCCATAAGTGGGGTTTCAAGCTATACGTTCTGTGCTCATTGTCTGGTTACGCTCACAACTTTGAAATTTATTCTGGAAAACAGAATATTCAATACTCAAGTGATGAACCTGATCTTGGTGTAGTAGGCAACACAGTCATCAGATTGTGCCGTTCAGTTCCACGAAGAATGAACCACATAATTTATTTTGATAATTTTTATTCGTCAATTCCTTTGTTATATTACCTACATACTCAAGGTATATATGCATTGGGCACAATTCAGAGGAACCGACTCGGTAAAACCTGTAAGTTGCCGGTAGTAAAAGAGTTTATGAAGGACTCTGTTCCAAGGGGATCATATGAGGAACTCGTAGCAGACTATGAAGGTGTGGATATCTCGGTAACTTGTTGGAAGGATAATAAGCTTGTTACGTTGGTATCTACTTATGTTGGTTCCGAACCGGCTGAAACTGTCAGCAGATTTGATAAGAAACAAAAGAAAAAAGTCTCTGTTGCCTGTCCAAAAATTGTAAAAGATTACAATGCCCACATGGGTGGAGTAGATTTGATGGACAGTAGCGACATCTGTCGAAAGAGCATGCGGAACTTCGATTAA
Protein
MSKRLAEQDIENILSVLENGDISEDEESIGDENDIDYYSNVQDLIQDLEDAEEDQYNANPDVPIEEDHLPDEFSVPGPSLSRPLSGVIGAAALRQRSRQLIWKKGNLEFSEDRISFTGNIDLSPEFAQLETPFQYFTLFLNDDILVKIVNETNLYITQKGISNCPPVNVKEIRQFLGIIIFMSVYHFPNVRSYWGKYGFKHIMETMTLNRFGKLRSVIHFNDNSLHKPVGHPNHDRLHKIRPVEAHLNKLFLSVAPFEQRLSLDEQMCSTKVAHFMKQYLPNKPHKWGFKLYVLCSLSGYAHNFEIYSGKQNIQYSSDEPDLGVVGNTVIRLCRSVPRRMNHIIYFDNFYSSIPLLYYLHTQGIYALGTIQRNRLGKTCKLPVVKEFMKDSVPRGSYEELVADYEGVDISVTCWKDNKLVTLVSTYVGSEPAETVSRFDKKQKKKVSVACPKIVKDYNAHMGGVDLMDSSDICRKSMRNFD
Summary
Uniprot
M4B148
S4UKH5
H9JXX7
A0A1A9VFM5
A0A212EHM2
J9M9C5
+ More
A0A2H8TNX6 T2MIT5 A0A2S2PBK5 J9LW60 X1X2F2 A0A2P8Y1P5 J9KQV2 A0A3B3I560 A0A3B5QXE4 A0A3B5R7A5 A0A146RLU8 A0A3B3IL37 A0A3P9HE00 T1HDP7 A0A3P9IAA1 A0A3B3H5M7 A0A146TTD4 A0A3B3I2Y1 A0A3P9I3M3 H2T2T7 A0A3P9JUS2 H2L551 A0A3S2N6X3 A0A3B1KA98 A0A3P8SS87 A0A3B3SQ04 A0A3B3TB08 A0A3B4ZMF2 A0A3Q1EXC1 A0A3N0XUJ7 A0A3B4Z7Z0 A0A3B3IBA3 A0A3B4EY15 A0A3B1KH96 J9L5C9 A0A3B3QLT9 A0A3P8ZBB8 A0A3B3S9X6 A0A087Y5J4 M3ZMJ7 A0A3B5N0U8 A0A3B3RJN9 A0A147BFV7 A0A3P9LFA7 A0A3B3HE36 A0A3P9NMX7 A0A3B3TU82 A0A146SI21 A0A146PY66 A0A315UTZ6 A0A3Q4MXL3 A0A3Q2QML2 A0A3B3XUL4 A0A3B3SQF8 A0A3B4D048 A0A3Q3E4B8 A0A3Q0RAX3 A0A212EM70 A0A3B3YQN0 A0A2A4JKE3 A0A3Q1JQK9 A0A3Q3E3V7 A0A3Q3G0X3 A0A3Q3NRD7 A0A3Q3E005 A0A3B4B8I5 A0A3Q3E559 A0A3Q3N2K1 A0A3Q3EH59 A0A3P9N476 A0A0S7I9G2 A0A3Q0SAC3 A0A3B5LTQ1 A0A3B4A161 A0A3B5LGD0 A0A0S7I592 A0A3Q4M2G3
A0A2H8TNX6 T2MIT5 A0A2S2PBK5 J9LW60 X1X2F2 A0A2P8Y1P5 J9KQV2 A0A3B3I560 A0A3B5QXE4 A0A3B5R7A5 A0A146RLU8 A0A3B3IL37 A0A3P9HE00 T1HDP7 A0A3P9IAA1 A0A3B3H5M7 A0A146TTD4 A0A3B3I2Y1 A0A3P9I3M3 H2T2T7 A0A3P9JUS2 H2L551 A0A3S2N6X3 A0A3B1KA98 A0A3P8SS87 A0A3B3SQ04 A0A3B3TB08 A0A3B4ZMF2 A0A3Q1EXC1 A0A3N0XUJ7 A0A3B4Z7Z0 A0A3B3IBA3 A0A3B4EY15 A0A3B1KH96 J9L5C9 A0A3B3QLT9 A0A3P8ZBB8 A0A3B3S9X6 A0A087Y5J4 M3ZMJ7 A0A3B5N0U8 A0A3B3RJN9 A0A147BFV7 A0A3P9LFA7 A0A3B3HE36 A0A3P9NMX7 A0A3B3TU82 A0A146SI21 A0A146PY66 A0A315UTZ6 A0A3Q4MXL3 A0A3Q2QML2 A0A3B3XUL4 A0A3B3SQF8 A0A3B4D048 A0A3Q3E4B8 A0A3Q0RAX3 A0A212EM70 A0A3B3YQN0 A0A2A4JKE3 A0A3Q1JQK9 A0A3Q3E3V7 A0A3Q3G0X3 A0A3Q3NRD7 A0A3Q3E005 A0A3B4B8I5 A0A3Q3E559 A0A3Q3N2K1 A0A3Q3EH59 A0A3P9N476 A0A0S7I9G2 A0A3Q0SAC3 A0A3B5LTQ1 A0A3B4A161 A0A3B5LGD0 A0A0S7I592 A0A3Q4M2G3
Pubmed
EMBL
BABH01034686
BABH01034687
BABH01034688
BABH01034689
JX392388
AGK25051.1
+ More
BABH01040586 BABH01040587 BABH01040588 AGBW02014856 OWR40986.1 ABLF02009142 GFXV01003527 MBW15332.1 HAAD01005618 CDG71850.1 GGMR01014224 MBY26843.1 ABLF02010159 ABLF02058363 ABLF02066746 PYGN01001043 PSN38182.1 ABLF02009898 GCES01116919 JAQ69403.1 ACPB03028099 GCES01090325 JAQ95997.1 CM012438 RVE75539.1 RJVU01062135 ROJ30485.1 ABLF02019811 ABLF02067232 ABLF02067266 AYCK01002766 GEGO01005727 JAR89677.1 GCES01106053 JAQ80269.1 GCES01137554 JAQ48768.1 NHOQ01002733 PWA15177.1 AGBW02013908 OWR42577.1 NWSH01001176 PCG72246.1 GBYX01432918 JAO48397.1 GBYX01432919 JAO48396.1
BABH01040586 BABH01040587 BABH01040588 AGBW02014856 OWR40986.1 ABLF02009142 GFXV01003527 MBW15332.1 HAAD01005618 CDG71850.1 GGMR01014224 MBY26843.1 ABLF02010159 ABLF02058363 ABLF02066746 PYGN01001043 PSN38182.1 ABLF02009898 GCES01116919 JAQ69403.1 ACPB03028099 GCES01090325 JAQ95997.1 CM012438 RVE75539.1 RJVU01062135 ROJ30485.1 ABLF02019811 ABLF02067232 ABLF02067266 AYCK01002766 GEGO01005727 JAR89677.1 GCES01106053 JAQ80269.1 GCES01137554 JAQ48768.1 NHOQ01002733 PWA15177.1 AGBW02013908 OWR42577.1 NWSH01001176 PCG72246.1 GBYX01432918 JAO48397.1 GBYX01432919 JAO48396.1
Proteomes
UP000005204
UP000078200
UP000007151
UP000007819
UP000245037
UP000001038
+ More
UP000002852 UP000265200 UP000015103 UP000005226 UP000018467 UP000265080 UP000261540 UP000261400 UP000257200 UP000261460 UP000265140 UP000028760 UP000261380 UP000265180 UP000242638 UP000261500 UP000265000 UP000261580 UP000261480 UP000261440 UP000261660 UP000261340 UP000218220 UP000265040 UP000261520
UP000002852 UP000265200 UP000015103 UP000005226 UP000018467 UP000265080 UP000261540 UP000261400 UP000257200 UP000261460 UP000265140 UP000028760 UP000261380 UP000265180 UP000242638 UP000261500 UP000265000 UP000261580 UP000261480 UP000261440 UP000261660 UP000261340 UP000218220 UP000265040 UP000261520
Interpro
IPR029526
PGBD
+ More
IPR042201 FH2_Formin_sf
IPR010472 FH3_dom
IPR015425 FH2_Formin
IPR011989 ARM-like
IPR014767 DAD_dom
IPR016024 ARM-type_fold
IPR010473 GTPase-bd
IPR014768 GBD/FH3_dom
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR013783 Ig-like_fold
IPR013151 Immunoglobulin
IPR042201 FH2_Formin_sf
IPR010472 FH3_dom
IPR015425 FH2_Formin
IPR011989 ARM-like
IPR014767 DAD_dom
IPR016024 ARM-type_fold
IPR010473 GTPase-bd
IPR014768 GBD/FH3_dom
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR007110 Ig-like_dom
IPR003599 Ig_sub
IPR013783 Ig-like_fold
IPR013151 Immunoglobulin
Gene 3D
ProteinModelPortal
M4B148
S4UKH5
H9JXX7
A0A1A9VFM5
A0A212EHM2
J9M9C5
+ More
A0A2H8TNX6 T2MIT5 A0A2S2PBK5 J9LW60 X1X2F2 A0A2P8Y1P5 J9KQV2 A0A3B3I560 A0A3B5QXE4 A0A3B5R7A5 A0A146RLU8 A0A3B3IL37 A0A3P9HE00 T1HDP7 A0A3P9IAA1 A0A3B3H5M7 A0A146TTD4 A0A3B3I2Y1 A0A3P9I3M3 H2T2T7 A0A3P9JUS2 H2L551 A0A3S2N6X3 A0A3B1KA98 A0A3P8SS87 A0A3B3SQ04 A0A3B3TB08 A0A3B4ZMF2 A0A3Q1EXC1 A0A3N0XUJ7 A0A3B4Z7Z0 A0A3B3IBA3 A0A3B4EY15 A0A3B1KH96 J9L5C9 A0A3B3QLT9 A0A3P8ZBB8 A0A3B3S9X6 A0A087Y5J4 M3ZMJ7 A0A3B5N0U8 A0A3B3RJN9 A0A147BFV7 A0A3P9LFA7 A0A3B3HE36 A0A3P9NMX7 A0A3B3TU82 A0A146SI21 A0A146PY66 A0A315UTZ6 A0A3Q4MXL3 A0A3Q2QML2 A0A3B3XUL4 A0A3B3SQF8 A0A3B4D048 A0A3Q3E4B8 A0A3Q0RAX3 A0A212EM70 A0A3B3YQN0 A0A2A4JKE3 A0A3Q1JQK9 A0A3Q3E3V7 A0A3Q3G0X3 A0A3Q3NRD7 A0A3Q3E005 A0A3B4B8I5 A0A3Q3E559 A0A3Q3N2K1 A0A3Q3EH59 A0A3P9N476 A0A0S7I9G2 A0A3Q0SAC3 A0A3B5LTQ1 A0A3B4A161 A0A3B5LGD0 A0A0S7I592 A0A3Q4M2G3
A0A2H8TNX6 T2MIT5 A0A2S2PBK5 J9LW60 X1X2F2 A0A2P8Y1P5 J9KQV2 A0A3B3I560 A0A3B5QXE4 A0A3B5R7A5 A0A146RLU8 A0A3B3IL37 A0A3P9HE00 T1HDP7 A0A3P9IAA1 A0A3B3H5M7 A0A146TTD4 A0A3B3I2Y1 A0A3P9I3M3 H2T2T7 A0A3P9JUS2 H2L551 A0A3S2N6X3 A0A3B1KA98 A0A3P8SS87 A0A3B3SQ04 A0A3B3TB08 A0A3B4ZMF2 A0A3Q1EXC1 A0A3N0XUJ7 A0A3B4Z7Z0 A0A3B3IBA3 A0A3B4EY15 A0A3B1KH96 J9L5C9 A0A3B3QLT9 A0A3P8ZBB8 A0A3B3S9X6 A0A087Y5J4 M3ZMJ7 A0A3B5N0U8 A0A3B3RJN9 A0A147BFV7 A0A3P9LFA7 A0A3B3HE36 A0A3P9NMX7 A0A3B3TU82 A0A146SI21 A0A146PY66 A0A315UTZ6 A0A3Q4MXL3 A0A3Q2QML2 A0A3B3XUL4 A0A3B3SQF8 A0A3B4D048 A0A3Q3E4B8 A0A3Q0RAX3 A0A212EM70 A0A3B3YQN0 A0A2A4JKE3 A0A3Q1JQK9 A0A3Q3E3V7 A0A3Q3G0X3 A0A3Q3NRD7 A0A3Q3E005 A0A3B4B8I5 A0A3Q3E559 A0A3Q3N2K1 A0A3Q3EH59 A0A3P9N476 A0A0S7I9G2 A0A3Q0SAC3 A0A3B5LTQ1 A0A3B4A161 A0A3B5LGD0 A0A0S7I592 A0A3Q4M2G3
Ontologies
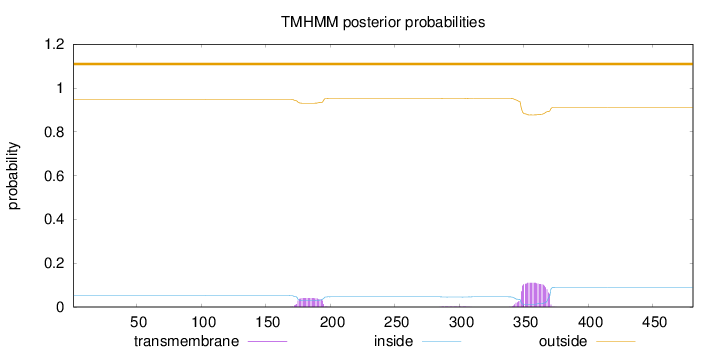
Topology
Length:
481
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.26363
Exp number, first 60 AAs:
0
Total prob of N-in:
0.05190
outside
1 - 481
Population Genetic Test Statistics
Pi
52.048112
Theta
15.838772
Tajima's D
-1.181131
CLR
110.31661
CSRT
0.106044697765112
Interpretation
Uncertain
