Gene
KWMTBOMO10285 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007267
Annotation
NADH_dehydrogenase_[ubiquinone]_1_beta_subcomplex_subunit_10_[Bombyx_mori]
Full name
NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10
Location in the cell
Cytoplasmic Reliability : 1.132 Nuclear Reliability : 1.312
Sequence
CDS
ATGGTACAAGACGGAAATCCTCCTGATGATAATGTGTTTAGAGCATTTTGCAATGCTCTCTACAATACAGTCGATGCTCCGGTAACATGGTTCCGAGAAACTGTTGTCGAACCAAACCAGAAGAAGTACCCTTGGTATCATCAAAACTACCGTCGTGTACCGACTATTGACCAGTGCTACGACGACGACGTCGTTTGCGACTTTGAGGCTAATGCTCAATTTAAACGAGACAGGGCAGTAGACTCTGAGATATTAAGCATTCTTAGGCAACGCTATGAAGATTGCATGATGTATGAGCAACCAGACCATGCAACTAAATGCAGGTCTCTTTGGGACAAGTACAAGTCTGCTGAGGAAGCTTGGTTCATTAAATATGGTGACCTCGGTGCTTATGGTGATGCGCGAAAAGCTTACATGAAGCAGAAGCATCGCATGGTCTGGGAACGACGCAATGGTCCCTTATCAGACCTTACAAAGTGA
Protein
MVQDGNPPDDNVFRAFCNALYNTVDAPVTWFRETVVEPNQKKYPWYHQNYRRVPTIDQCYDDDVVCDFEANAQFKRDRAVDSEILSILRQRYEDCMMYEQPDHATKCRSLWDKYKSAEEAWFIKYGDLGAYGDARKAYMKQKHRMVWERRNGPLSDLTK
Summary
Description
Accessory subunit of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I), that is believed not to be involved in catalysis. Complex I functions in the transfer of electrons from NADH to the respiratory chain. The immediate electron acceptor for the enzyme is believed to be ubiquinone (By similarity).
Subunit
Complex I is composed of 45 different subunits.
Similarity
Belongs to the complex I NDUFB10 subunit family.
Keywords
Complete proteome
Direct protein sequencing
Electron transport
Membrane
Mitochondrion
Mitochondrion inner membrane
Reference proteome
Respiratory chain
Transport
Feature
chain NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10
Uniprot
Q1HPL8
S4PWZ7
A0A212EM25
A0A194QX41
A0A194QH76
A0A2H1WLI3
+ More
A0A1E1W6L6 A0A2A4JHN4 A0A1A9WKT1 A0A182LUK1 A0A182QR70 B0WCI6 A0A1A9ZDC7 A0A182K359 A0A182MZA3 Q7Q3A0 Q17JZ6 A0A023EHR6 A0A182X1G9 A0A182VC00 A0A182L2L6 A0A182IEE9 A0A023EHN2 A0A1Q3FJ54 T1E870 A0A2M3Z5E6 A0A2M4AAW3 A0A2M4CZ70 A0A182PJQ0 T1PBS2 A0A0K8TNN0 T1E348 A0A1L8EEY8 A0A0L0C689 A0A0M5J1G1 A0A0M4E3X1 A0A1L8E0K2 A0A1B0CKK0 A0A1L8E0F1 A0A1I8PIH3 A0A1B0F9N9 A0A1A9XL36 A0A1B0B7Q3 A0A1A9V9T0 A0A1J1I0F5 B4NXX1 B4INK6 A0AQ35 Q9VQR2 W8BLU7 A0AQ24 B3MP23 A0A3B0JA55 B4MU32 A0A336MMU1 A0A0P4VN49 R4G7W4 A0A034WKY3 Q29MC0 B4G9G5 A0A1W4UU52 A0A0A9YKT9 B4KHH7 B3N326 B4LTX8 B4JBH3 C4N167 A0A0A1XCY9 D6WWB4 A0A023F817 A0A0K8VBB3 A0A0V0G624 A0A069DPK6 A0A2R7X1Q7 A0A1Y1MRR1 A0A1B6DSM6 A0A067RIY0 U5EWT9 N6UB84 J3JZ58 A0A0P4WJ44 A0A3R7MJN5 A0A1W4WUT3 B4Q9K7 A0A1B0DNQ8 A0A0L7R8R0 V9IMC1 A0A2A3EDM5 T1J1T5 E2BMC3 A0A1B6M7K7 A0A3L8DM95 E2AWW2 A0A087ZXL3 A0A0A1CMK0 A0A151JUE8 A0A1B6JN66 A0A026W0K4 A0A195DL42 A0A232EY78
A0A1E1W6L6 A0A2A4JHN4 A0A1A9WKT1 A0A182LUK1 A0A182QR70 B0WCI6 A0A1A9ZDC7 A0A182K359 A0A182MZA3 Q7Q3A0 Q17JZ6 A0A023EHR6 A0A182X1G9 A0A182VC00 A0A182L2L6 A0A182IEE9 A0A023EHN2 A0A1Q3FJ54 T1E870 A0A2M3Z5E6 A0A2M4AAW3 A0A2M4CZ70 A0A182PJQ0 T1PBS2 A0A0K8TNN0 T1E348 A0A1L8EEY8 A0A0L0C689 A0A0M5J1G1 A0A0M4E3X1 A0A1L8E0K2 A0A1B0CKK0 A0A1L8E0F1 A0A1I8PIH3 A0A1B0F9N9 A0A1A9XL36 A0A1B0B7Q3 A0A1A9V9T0 A0A1J1I0F5 B4NXX1 B4INK6 A0AQ35 Q9VQR2 W8BLU7 A0AQ24 B3MP23 A0A3B0JA55 B4MU32 A0A336MMU1 A0A0P4VN49 R4G7W4 A0A034WKY3 Q29MC0 B4G9G5 A0A1W4UU52 A0A0A9YKT9 B4KHH7 B3N326 B4LTX8 B4JBH3 C4N167 A0A0A1XCY9 D6WWB4 A0A023F817 A0A0K8VBB3 A0A0V0G624 A0A069DPK6 A0A2R7X1Q7 A0A1Y1MRR1 A0A1B6DSM6 A0A067RIY0 U5EWT9 N6UB84 J3JZ58 A0A0P4WJ44 A0A3R7MJN5 A0A1W4WUT3 B4Q9K7 A0A1B0DNQ8 A0A0L7R8R0 V9IMC1 A0A2A3EDM5 T1J1T5 E2BMC3 A0A1B6M7K7 A0A3L8DM95 E2AWW2 A0A087ZXL3 A0A0A1CMK0 A0A151JUE8 A0A1B6JN66 A0A026W0K4 A0A195DL42 A0A232EY78
Pubmed
11280994
23622113
22118469
26354079
12364791
14747013
+ More
17210077 17510324 24945155 20966253 25315136 26369729 24330624 26108605 17994087 17550304 16951084 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19126864 26109357 26109356 24495485 18057021 27129103 25348373 15632085 25401762 26823975 19576987 25830018 18362917 19820115 25474469 26334808 28004739 24845553 23537049 22516182 20798317 30249741 24508170 28648823
17210077 17510324 24945155 20966253 25315136 26369729 24330624 26108605 17994087 17550304 16951084 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 19126864 26109357 26109356 24495485 18057021 27129103 25348373 15632085 25401762 26823975 19576987 25830018 18362917 19820115 25474469 26334808 28004739 24845553 23537049 22516182 20798317 30249741 24508170 28648823
EMBL
DQ443384
ABF51473.1
GAIX01005918
JAA86642.1
AGBW02013944
OWR42533.1
+ More
KQ461155 KPJ08141.1 KQ459232 KPJ02796.1 ODYU01009447 SOQ53888.1 GDQN01008449 JAT82605.1 NWSH01001441 PCG71228.1 AXCM01003708 AXCM01003709 AXCN02002070 DS231888 EDS43557.1 AAAB01008964 EAA12342.3 CH477229 EAT47004.1 GAPW01005204 JAC08394.1 APCN01005201 GAPW01005203 JAC08395.1 GFDL01007428 JAV27617.1 GAMD01002432 JAA99158.1 GGFM01002985 MBW23736.1 GGFK01004603 MBW37924.1 GGFL01006323 MBW70501.1 KA645560 AFP60189.1 GDAI01001606 JAI15997.1 GALA01000695 JAA94157.1 GFDG01001696 JAV17103.1 JRES01000842 KNC27800.1 CP012523 ALC38968.1 ALC38408.1 GFDF01001875 JAV12209.1 AJWK01016343 GFDF01001874 JAV12210.1 CCAG010005376 JXJN01009699 CVRI01000037 CRK93675.1 CM000157 EDW87542.1 KRJ97203.1 CH676463 EDW44615.1 AM294773 CM002910 CAL26759.1 KMY87896.1 AE014134 AY060865 AY113547 AM294761 AM294763 AM294767 AM294769 AM294772 FM245890 FM245891 FM245892 FM245894 FM245895 FM245896 FM245897 FM245898 AAF51103.1 AAL28413.1 AAM29552.1 AAN10369.1 CAL26747.1 CAL26749.1 CAL26753.1 CAL26755.1 CAL26758.1 CAR93816.1 CAR93817.1 CAR93818.1 CAR93820.1 CAR93821.1 CAR93822.1 CAR93823.1 CAR93824.1 GAMC01004495 JAC02061.1 AM294762 AM294764 AM294765 AM294766 AM294768 AM294770 AM294771 FM245893 CAL26748.1 CAL26750.1 CAL26751.1 CAL26752.1 CAL26754.1 CAL26756.1 CAL26757.1 CAR93819.1 CH902620 EDV31189.1 KPU73258.1 OUUW01000004 SPP79174.1 CH963852 EDW75621.1 UFQT01001740 UFQT01003943 SSX31606.1 SSX35382.1 GDKW01002286 JAI54309.1 ACPB03011663 GAHY01002017 JAA75493.1 GAKP01004509 JAC54443.1 CH379060 EAL33774.3 CH479180 EDW28995.1 GBHO01011358 GBRD01006410 GDHC01005355 JAG32246.1 JAG59411.1 JAQ13274.1 CH933807 EDW11241.1 CH954177 EDV57625.1 KQS70045.1 CH940649 EDW64029.1 KRF81396.1 CH916368 EDW03996.1 EZ048840 ACN69132.1 GBXI01016676 GBXI01005964 JAC97615.1 JAD08328.1 KQ971361 EFA09231.1 GBBI01001307 JAC17405.1 GDHF01021869 GDHF01016504 GDHF01011136 GDHF01001639 JAI30445.1 JAI35810.1 JAI41178.1 JAI50675.1 GECL01003309 JAP02815.1 GBGD01003268 JAC85621.1 KK856086 PTY24825.1 GEZM01023426 JAV88392.1 GEDC01008635 JAS28663.1 KK852478 KDR22988.1 GANO01002817 JAB57054.1 APGK01035705 APGK01035706 KB740928 KB632428 ENN77916.1 ERL95416.1 BT128539 AEE63496.1 GDRN01042869 JAI67011.1 QCYY01000534 ROT84484.1 CM000361 EDX03618.1 AJVK01017678 KQ414631 KOC67229.1 JR052564 AEY61726.1 KZ288288 PBC29329.1 JH431789 GL449185 EFN83152.1 GEBQ01008060 JAT31917.1 QOIP01000006 RLU21446.1 GL443425 EFN62082.1 KM275727 KM275757 AIX97483.1 AIX97513.1 KQ981745 KYN36187.1 GECU01006995 JAT00712.1 KK107503 EZA49570.1 KQ980762 KYN13566.1 NNAY01001707 OXU23168.1
KQ461155 KPJ08141.1 KQ459232 KPJ02796.1 ODYU01009447 SOQ53888.1 GDQN01008449 JAT82605.1 NWSH01001441 PCG71228.1 AXCM01003708 AXCM01003709 AXCN02002070 DS231888 EDS43557.1 AAAB01008964 EAA12342.3 CH477229 EAT47004.1 GAPW01005204 JAC08394.1 APCN01005201 GAPW01005203 JAC08395.1 GFDL01007428 JAV27617.1 GAMD01002432 JAA99158.1 GGFM01002985 MBW23736.1 GGFK01004603 MBW37924.1 GGFL01006323 MBW70501.1 KA645560 AFP60189.1 GDAI01001606 JAI15997.1 GALA01000695 JAA94157.1 GFDG01001696 JAV17103.1 JRES01000842 KNC27800.1 CP012523 ALC38968.1 ALC38408.1 GFDF01001875 JAV12209.1 AJWK01016343 GFDF01001874 JAV12210.1 CCAG010005376 JXJN01009699 CVRI01000037 CRK93675.1 CM000157 EDW87542.1 KRJ97203.1 CH676463 EDW44615.1 AM294773 CM002910 CAL26759.1 KMY87896.1 AE014134 AY060865 AY113547 AM294761 AM294763 AM294767 AM294769 AM294772 FM245890 FM245891 FM245892 FM245894 FM245895 FM245896 FM245897 FM245898 AAF51103.1 AAL28413.1 AAM29552.1 AAN10369.1 CAL26747.1 CAL26749.1 CAL26753.1 CAL26755.1 CAL26758.1 CAR93816.1 CAR93817.1 CAR93818.1 CAR93820.1 CAR93821.1 CAR93822.1 CAR93823.1 CAR93824.1 GAMC01004495 JAC02061.1 AM294762 AM294764 AM294765 AM294766 AM294768 AM294770 AM294771 FM245893 CAL26748.1 CAL26750.1 CAL26751.1 CAL26752.1 CAL26754.1 CAL26756.1 CAL26757.1 CAR93819.1 CH902620 EDV31189.1 KPU73258.1 OUUW01000004 SPP79174.1 CH963852 EDW75621.1 UFQT01001740 UFQT01003943 SSX31606.1 SSX35382.1 GDKW01002286 JAI54309.1 ACPB03011663 GAHY01002017 JAA75493.1 GAKP01004509 JAC54443.1 CH379060 EAL33774.3 CH479180 EDW28995.1 GBHO01011358 GBRD01006410 GDHC01005355 JAG32246.1 JAG59411.1 JAQ13274.1 CH933807 EDW11241.1 CH954177 EDV57625.1 KQS70045.1 CH940649 EDW64029.1 KRF81396.1 CH916368 EDW03996.1 EZ048840 ACN69132.1 GBXI01016676 GBXI01005964 JAC97615.1 JAD08328.1 KQ971361 EFA09231.1 GBBI01001307 JAC17405.1 GDHF01021869 GDHF01016504 GDHF01011136 GDHF01001639 JAI30445.1 JAI35810.1 JAI41178.1 JAI50675.1 GECL01003309 JAP02815.1 GBGD01003268 JAC85621.1 KK856086 PTY24825.1 GEZM01023426 JAV88392.1 GEDC01008635 JAS28663.1 KK852478 KDR22988.1 GANO01002817 JAB57054.1 APGK01035705 APGK01035706 KB740928 KB632428 ENN77916.1 ERL95416.1 BT128539 AEE63496.1 GDRN01042869 JAI67011.1 QCYY01000534 ROT84484.1 CM000361 EDX03618.1 AJVK01017678 KQ414631 KOC67229.1 JR052564 AEY61726.1 KZ288288 PBC29329.1 JH431789 GL449185 EFN83152.1 GEBQ01008060 JAT31917.1 QOIP01000006 RLU21446.1 GL443425 EFN62082.1 KM275727 KM275757 AIX97483.1 AIX97513.1 KQ981745 KYN36187.1 GECU01006995 JAT00712.1 KK107503 EZA49570.1 KQ980762 KYN13566.1 NNAY01001707 OXU23168.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000053268
UP000218220
UP000091820
+ More
UP000075883 UP000075886 UP000002320 UP000092445 UP000075881 UP000075884 UP000007062 UP000008820 UP000076407 UP000075903 UP000075882 UP000075840 UP000075885 UP000095301 UP000037069 UP000092553 UP000092461 UP000095300 UP000092444 UP000092443 UP000092460 UP000078200 UP000183832 UP000002282 UP000001292 UP000000803 UP000007801 UP000268350 UP000007798 UP000015103 UP000001819 UP000008744 UP000192221 UP000009192 UP000008711 UP000008792 UP000001070 UP000007266 UP000027135 UP000019118 UP000030742 UP000283509 UP000192223 UP000000304 UP000092462 UP000053825 UP000242457 UP000008237 UP000279307 UP000000311 UP000005203 UP000078541 UP000053097 UP000078492 UP000215335
UP000075883 UP000075886 UP000002320 UP000092445 UP000075881 UP000075884 UP000007062 UP000008820 UP000076407 UP000075903 UP000075882 UP000075840 UP000075885 UP000095301 UP000037069 UP000092553 UP000092461 UP000095300 UP000092444 UP000092443 UP000092460 UP000078200 UP000183832 UP000002282 UP000001292 UP000000803 UP000007801 UP000268350 UP000007798 UP000015103 UP000001819 UP000008744 UP000192221 UP000009192 UP000008711 UP000008792 UP000001070 UP000007266 UP000027135 UP000019118 UP000030742 UP000283509 UP000192223 UP000000304 UP000092462 UP000053825 UP000242457 UP000008237 UP000279307 UP000000311 UP000005203 UP000078541 UP000053097 UP000078492 UP000215335
Interpro
ProteinModelPortal
Q1HPL8
S4PWZ7
A0A212EM25
A0A194QX41
A0A194QH76
A0A2H1WLI3
+ More
A0A1E1W6L6 A0A2A4JHN4 A0A1A9WKT1 A0A182LUK1 A0A182QR70 B0WCI6 A0A1A9ZDC7 A0A182K359 A0A182MZA3 Q7Q3A0 Q17JZ6 A0A023EHR6 A0A182X1G9 A0A182VC00 A0A182L2L6 A0A182IEE9 A0A023EHN2 A0A1Q3FJ54 T1E870 A0A2M3Z5E6 A0A2M4AAW3 A0A2M4CZ70 A0A182PJQ0 T1PBS2 A0A0K8TNN0 T1E348 A0A1L8EEY8 A0A0L0C689 A0A0M5J1G1 A0A0M4E3X1 A0A1L8E0K2 A0A1B0CKK0 A0A1L8E0F1 A0A1I8PIH3 A0A1B0F9N9 A0A1A9XL36 A0A1B0B7Q3 A0A1A9V9T0 A0A1J1I0F5 B4NXX1 B4INK6 A0AQ35 Q9VQR2 W8BLU7 A0AQ24 B3MP23 A0A3B0JA55 B4MU32 A0A336MMU1 A0A0P4VN49 R4G7W4 A0A034WKY3 Q29MC0 B4G9G5 A0A1W4UU52 A0A0A9YKT9 B4KHH7 B3N326 B4LTX8 B4JBH3 C4N167 A0A0A1XCY9 D6WWB4 A0A023F817 A0A0K8VBB3 A0A0V0G624 A0A069DPK6 A0A2R7X1Q7 A0A1Y1MRR1 A0A1B6DSM6 A0A067RIY0 U5EWT9 N6UB84 J3JZ58 A0A0P4WJ44 A0A3R7MJN5 A0A1W4WUT3 B4Q9K7 A0A1B0DNQ8 A0A0L7R8R0 V9IMC1 A0A2A3EDM5 T1J1T5 E2BMC3 A0A1B6M7K7 A0A3L8DM95 E2AWW2 A0A087ZXL3 A0A0A1CMK0 A0A151JUE8 A0A1B6JN66 A0A026W0K4 A0A195DL42 A0A232EY78
A0A1E1W6L6 A0A2A4JHN4 A0A1A9WKT1 A0A182LUK1 A0A182QR70 B0WCI6 A0A1A9ZDC7 A0A182K359 A0A182MZA3 Q7Q3A0 Q17JZ6 A0A023EHR6 A0A182X1G9 A0A182VC00 A0A182L2L6 A0A182IEE9 A0A023EHN2 A0A1Q3FJ54 T1E870 A0A2M3Z5E6 A0A2M4AAW3 A0A2M4CZ70 A0A182PJQ0 T1PBS2 A0A0K8TNN0 T1E348 A0A1L8EEY8 A0A0L0C689 A0A0M5J1G1 A0A0M4E3X1 A0A1L8E0K2 A0A1B0CKK0 A0A1L8E0F1 A0A1I8PIH3 A0A1B0F9N9 A0A1A9XL36 A0A1B0B7Q3 A0A1A9V9T0 A0A1J1I0F5 B4NXX1 B4INK6 A0AQ35 Q9VQR2 W8BLU7 A0AQ24 B3MP23 A0A3B0JA55 B4MU32 A0A336MMU1 A0A0P4VN49 R4G7W4 A0A034WKY3 Q29MC0 B4G9G5 A0A1W4UU52 A0A0A9YKT9 B4KHH7 B3N326 B4LTX8 B4JBH3 C4N167 A0A0A1XCY9 D6WWB4 A0A023F817 A0A0K8VBB3 A0A0V0G624 A0A069DPK6 A0A2R7X1Q7 A0A1Y1MRR1 A0A1B6DSM6 A0A067RIY0 U5EWT9 N6UB84 J3JZ58 A0A0P4WJ44 A0A3R7MJN5 A0A1W4WUT3 B4Q9K7 A0A1B0DNQ8 A0A0L7R8R0 V9IMC1 A0A2A3EDM5 T1J1T5 E2BMC3 A0A1B6M7K7 A0A3L8DM95 E2AWW2 A0A087ZXL3 A0A0A1CMK0 A0A151JUE8 A0A1B6JN66 A0A026W0K4 A0A195DL42 A0A232EY78
PDB
5LNK
E-value=1.07763e-20,
Score=241
Ontologies
PATHWAY
GO
PANTHER
Topology
Subcellular location
Mitochondrion inner membrane
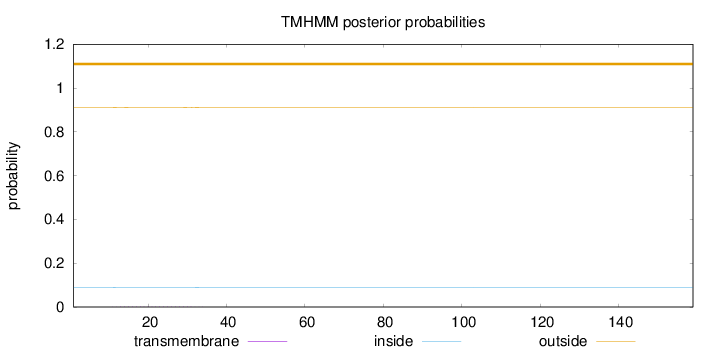
Length:
159
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03287
Exp number, first 60 AAs:
0.03248
Total prob of N-in:
0.08896
outside
1 - 159
Population Genetic Test Statistics
Pi
27.096339
Theta
39.734007
Tajima's D
-0.8329
CLR
116.550675
CSRT
0.166741662916854
Interpretation
Uncertain
