Gene
KWMTBOMO10283 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007268
Annotation
PREDICTED:_proteasome_26S_non-ATPase_subunit_12_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.355
Sequence
CDS
ATGACTACAAACGGAGATATCGGCGGTCTCGATGCTAGTGGAAAAATAATAAAAATGGAAGTGGACTATAGTGCAACATGTGATGAGAAACTGCCCTTGTGGAAATCATGGGCAGCCCAAGGCAAAATTCAAGAAGCTATCGATCAGTTATTGGCTCTTGAAAAACAGACTAGGACTGGTGCCGATATGGTTTCCACTTCAAGAATCCTAGTGACAGTGGTTCAGATATATTTTGAAGCTAAGAACTGGTCTGCTCTCAATGATCATATAGTTGTTTTGTCCAAAAGAAGATCCCAATTAAAACAAGCTGTTGTTAAAATGGTTCAGGAATGCTACACTTATGTAGACAAAACACCCGATAAAGAAACAAAGATAAAACTAATTGAAACATTGAGAACCATAACAGAAGGAAAGATCTATGTAGAAGTTGAAAGAGCGAGACTGACTCATATTTTAGCCAAAATTCGAGAAGAAGAAGGCAATGTAGCAGAGGCTGCAAAAATAATACAAGAACTTCAAGTAGAGACATATGGTTCAATGGACAAGAGGGAAAAAGTTGAGTTAATTCTGGAACAGATGAGATTGTGTCTTGCTATCAAAGATTATGTCCGTACACAGATCATATCAAAGAAAATCAATACAAAATTTTTTGAAGACGAAAATACTCAGGAGCTGAAAGAGAAGTTTTACCGACTGATGATAGCTGTGGACCAGCATAATGGACAATATCTGTCAGTCTGTCGGCATTTCCGAGCTTTAGGAACTGCTGGGGGCCCTGAAGCACTTATTGGCAGTGTTGTATTCCTTATATTGGCACCATATGACAATGAACAGGCAGATCTCACACATAGAGTAAATGAAGATAAGGAGTTGGATAAACTTCCTGATTATAAAGAATTGTTACGTCTATTCATAAATCCTGAGATCATCAGATGGAACACTCTATGTTCTTCTTACGAGAAGATGCTTCGGGCCACACCGTACTTTGACGCGTCAGACGACAAGGGTCAAGAACGCTGGAACGATCTTAAGAACAGAGTTGTTGAACATAATATTCGCATAATGTCAATGTACTATACTCGCATCACGCTCAAACGTATGAGTGAACTTCTTGGTCTGAGCGAGACGGAAACAGAAGAGGCCCTGAGTCAGCTTGTTGTGAGTGCAGTTGTTAAAGCCAAGATCGATCGTCCAGCTGGCGTTGTACACTTTAGCCTGAATATGGACGCGTCAGATCGTCTCAACGAATGGTCGAACAATCTGAACACATTGATGCAACTTGTCAATAAAACAACACATCTCATTAACAAAGAGGAATGCGTTCACAAACATTTGTTGGCTACTGCAGAGTAA
Protein
MTTNGDIGGLDASGKIIKMEVDYSATCDEKLPLWKSWAAQGKIQEAIDQLLALEKQTRTGADMVSTSRILVTVVQIYFEAKNWSALNDHIVVLSKRRSQLKQAVVKMVQECYTYVDKTPDKETKIKLIETLRTITEGKIYVEVERARLTHILAKIREEEGNVAEAAKIIQELQVETYGSMDKREKVELILEQMRLCLAIKDYVRTQIISKKINTKFFEDENTQELKEKFYRLMIAVDQHNGQYLSVCRHFRALGTAGGPEALIGSVVFLILAPYDNEQADLTHRVNEDKELDKLPDYKELLRLFINPEIIRWNTLCSSYEKMLRATPYFDASDDKGQERWNDLKNRVVEHNIRIMSMYYTRITLKRMSELLGLSETETEEALSQLVVSAVVKAKIDRPAGVVHFSLNMDASDRLNEWSNNLNTLMQLVNKTTHLINKEECVHKHLLATAE
Summary
Uniprot
H9JCM2
Q2F5W8
A0A3S2TFP1
A0A2A4IZ53
A0A2H1WLC9
S4P9I7
+ More
I4DMA9 A0A194QS44 A0A194QBK9 D6WVQ2 A0A0T6B2B6 A0A1W4WEY9 A0A2J7PKP9 A0A067RIG7 A0A1B6ER76 A0A2P8XQ56 A0A1Y1MDM2 E0VNS1 N6TS51 U4TWV0 A0A0V0G3D9 A0A023F967 A0A069DZ24 A0A1S4E7U6 A0A224XP92 A0A0A9ZGG0 A0A0P4VSA9 R4G476 A0A2R7WXR0 A0A087UIR5 A0A0C5B0H1 A0A2R5LLC6 V5IA60 J9JUQ2 A0A131Z0W6 L7LZY7 A0A0C9PMI1 A0A171BB79 W5N823 E2AI04 T1J1H9 A0A131XDL0 A0A3L8DIS9 E2BW38 A0A3P9JTW7 A0A3P9LHH5 A0A1Z5KWS0 A0A131XVD4 A0A3B3RUB1 A0A0S7G0W9 A0A3B3DB81 A0A2B4SU00 A0A3M6UQ20 A0A3Q2YD84 A0A0B8RWM7 V8NWQ4 E9GVL8 A0A3Q3VQH3 A0A3Q3GI97 A0A3B3HKL0 A0A1A7WW76 I3JH38 A0A164ZVQ4 A0A3Q2GEL9 A0A2I4BWH2 A0A3P9MSX1 A0A1A8J2H2 A0A1A8DY44 A0A1A8G6P6 A0A1A8ARA8 A0A3Q1HZ04 A0A087Y5S0 A0A1A8QZC0 A0A1A8L461 A0A0N7ZJY8 A0A0P6I624 G3Q3S8 A0A3B3UV04 E9IJF3 A0A3Q3MFH1 H3CHW8 A0A0F7Z0Z0 A0A2R2YUM9 A0A3S2PH67 A0A3B5QXX0 A0A3B5AGZ2 A0A146MTG9 A0A3B4UB02 Q4T0S8 W5KWC5 A0A3B4DDI3 A0A3P8S093 A0A0P7UVL0 A0A3B5KFX6 A0A3Q3F8P0 A0A2U9BP45 A0A232FK97 A0A3Q1FXJ6 C3Z7L1 K1QQK1
I4DMA9 A0A194QS44 A0A194QBK9 D6WVQ2 A0A0T6B2B6 A0A1W4WEY9 A0A2J7PKP9 A0A067RIG7 A0A1B6ER76 A0A2P8XQ56 A0A1Y1MDM2 E0VNS1 N6TS51 U4TWV0 A0A0V0G3D9 A0A023F967 A0A069DZ24 A0A1S4E7U6 A0A224XP92 A0A0A9ZGG0 A0A0P4VSA9 R4G476 A0A2R7WXR0 A0A087UIR5 A0A0C5B0H1 A0A2R5LLC6 V5IA60 J9JUQ2 A0A131Z0W6 L7LZY7 A0A0C9PMI1 A0A171BB79 W5N823 E2AI04 T1J1H9 A0A131XDL0 A0A3L8DIS9 E2BW38 A0A3P9JTW7 A0A3P9LHH5 A0A1Z5KWS0 A0A131XVD4 A0A3B3RUB1 A0A0S7G0W9 A0A3B3DB81 A0A2B4SU00 A0A3M6UQ20 A0A3Q2YD84 A0A0B8RWM7 V8NWQ4 E9GVL8 A0A3Q3VQH3 A0A3Q3GI97 A0A3B3HKL0 A0A1A7WW76 I3JH38 A0A164ZVQ4 A0A3Q2GEL9 A0A2I4BWH2 A0A3P9MSX1 A0A1A8J2H2 A0A1A8DY44 A0A1A8G6P6 A0A1A8ARA8 A0A3Q1HZ04 A0A087Y5S0 A0A1A8QZC0 A0A1A8L461 A0A0N7ZJY8 A0A0P6I624 G3Q3S8 A0A3B3UV04 E9IJF3 A0A3Q3MFH1 H3CHW8 A0A0F7Z0Z0 A0A2R2YUM9 A0A3S2PH67 A0A3B5QXX0 A0A3B5AGZ2 A0A146MTG9 A0A3B4UB02 Q4T0S8 W5KWC5 A0A3B4DDI3 A0A3P8S093 A0A0P7UVL0 A0A3B5KFX6 A0A3Q3F8P0 A0A2U9BP45 A0A232FK97 A0A3Q1FXJ6 C3Z7L1 K1QQK1
Pubmed
19121390
23622113
22651552
26354079
18362917
19820115
+ More
24845553 29403074 28004739 20566863 23537049 25474469 26334808 25401762 26823975 27129103 25653432 26830274 25576852 20798317 28049606 30249741 17554307 28528879 29652888 29240929 29451363 30382153 25476704 24297900 21292972 25186727 21282665 15496914 23542700 25329095 21551351 28648823 18563158 22992520
24845553 29403074 28004739 20566863 23537049 25474469 26334808 25401762 26823975 27129103 25653432 26830274 25576852 20798317 28049606 30249741 17554307 28528879 29652888 29240929 29451363 30382153 25476704 24297900 21292972 25186727 21282665 15496914 23542700 25329095 21551351 28648823 18563158 22992520
EMBL
BABH01034655
DQ311305
ABD36249.1
RSAL01000189
RVE44734.1
NWSH01004502
+ More
PCG65041.1 ODYU01009447 SOQ53885.1 GAIX01003739 JAA88821.1 AK402427 BAM19049.1 KQ461155 KPJ08139.1 KQ459232 KPJ02799.1 KQ971358 EFA08601.1 LJIG01016203 KRT81307.1 NEVH01024535 PNF16906.1 KK852663 KDR19074.1 GECZ01029351 JAS40418.1 PYGN01001553 PSN34115.1 GEZM01036699 JAV82415.1 DS235346 EEB15027.1 APGK01056791 KB741277 ENN71126.1 KB631786 ERL86079.1 GECL01003539 JAP02585.1 GBBI01000670 JAC18042.1 GBGD01001385 JAC87504.1 GFTR01006443 JAW09983.1 GBHO01002539 GBRD01014104 GDHC01010693 JAG41065.1 JAG51722.1 JAQ07936.1 GDKW01001254 JAI55341.1 ACPB03005390 GAHY01001021 JAA76489.1 KK855644 PTY23395.1 KK119981 KFM77254.1 KM999969 AJK90584.1 GGLE01006200 MBY10326.1 GALX01001750 JAB66716.1 ABLF02036253 GEDV01004182 JAP84375.1 GACK01007413 JAA57621.1 GBYB01002293 JAG72060.1 GEMB01000041 JAS03068.1 AHAT01000495 GL439621 EFN66912.1 JH431785 GEFH01004393 JAP64188.1 QOIP01000007 RLU20350.1 GL451091 EFN80070.1 GFJQ02007431 JAV99538.1 GEFM01004542 GEGO01001134 JAP71254.1 JAR94270.1 GBYX01459740 JAO21819.1 LSMT01000028 PFX32032.1 RCHS01001049 RMX55448.1 GBSH01002134 JAG66892.1 AZIM01001571 ETE66401.1 GL732568 EFX76433.1 HADW01008573 HADX01001392 SBP09973.1 AERX01050221 LRGB01000687 KZS16839.1 HAED01017256 HAEE01014724 SBR03701.1 HADZ01009847 HAEA01010372 SBQ38852.1 HAEB01020350 HAEC01005908 SBQ66877.1 HADY01019074 HAEJ01013570 SBP57559.1 AYCK01011808 AYCK01011809 HAEI01006736 HAEH01017554 SBR98857.1 HAEF01002040 HAEG01015735 SBR39422.1 GDIP01238033 JAI85368.1 GDIQ01025965 JAN68772.1 GL763766 EFZ19304.1 GBEW01000762 JAI09603.1 KM522790 AKN80416.1 CM012437 RVE75679.1 GCES01163359 JAQ22963.1 CAAE01010875 CAF93504.1 JARO02000460 KPP78479.1 CP026250 AWP05907.1 NNAY01000124 OXU30747.1 GG666591 EEN51366.1 JH817356 EKC39252.1
PCG65041.1 ODYU01009447 SOQ53885.1 GAIX01003739 JAA88821.1 AK402427 BAM19049.1 KQ461155 KPJ08139.1 KQ459232 KPJ02799.1 KQ971358 EFA08601.1 LJIG01016203 KRT81307.1 NEVH01024535 PNF16906.1 KK852663 KDR19074.1 GECZ01029351 JAS40418.1 PYGN01001553 PSN34115.1 GEZM01036699 JAV82415.1 DS235346 EEB15027.1 APGK01056791 KB741277 ENN71126.1 KB631786 ERL86079.1 GECL01003539 JAP02585.1 GBBI01000670 JAC18042.1 GBGD01001385 JAC87504.1 GFTR01006443 JAW09983.1 GBHO01002539 GBRD01014104 GDHC01010693 JAG41065.1 JAG51722.1 JAQ07936.1 GDKW01001254 JAI55341.1 ACPB03005390 GAHY01001021 JAA76489.1 KK855644 PTY23395.1 KK119981 KFM77254.1 KM999969 AJK90584.1 GGLE01006200 MBY10326.1 GALX01001750 JAB66716.1 ABLF02036253 GEDV01004182 JAP84375.1 GACK01007413 JAA57621.1 GBYB01002293 JAG72060.1 GEMB01000041 JAS03068.1 AHAT01000495 GL439621 EFN66912.1 JH431785 GEFH01004393 JAP64188.1 QOIP01000007 RLU20350.1 GL451091 EFN80070.1 GFJQ02007431 JAV99538.1 GEFM01004542 GEGO01001134 JAP71254.1 JAR94270.1 GBYX01459740 JAO21819.1 LSMT01000028 PFX32032.1 RCHS01001049 RMX55448.1 GBSH01002134 JAG66892.1 AZIM01001571 ETE66401.1 GL732568 EFX76433.1 HADW01008573 HADX01001392 SBP09973.1 AERX01050221 LRGB01000687 KZS16839.1 HAED01017256 HAEE01014724 SBR03701.1 HADZ01009847 HAEA01010372 SBQ38852.1 HAEB01020350 HAEC01005908 SBQ66877.1 HADY01019074 HAEJ01013570 SBP57559.1 AYCK01011808 AYCK01011809 HAEI01006736 HAEH01017554 SBR98857.1 HAEF01002040 HAEG01015735 SBR39422.1 GDIP01238033 JAI85368.1 GDIQ01025965 JAN68772.1 GL763766 EFZ19304.1 GBEW01000762 JAI09603.1 KM522790 AKN80416.1 CM012437 RVE75679.1 GCES01163359 JAQ22963.1 CAAE01010875 CAF93504.1 JARO02000460 KPP78479.1 CP026250 AWP05907.1 NNAY01000124 OXU30747.1 GG666591 EEN51366.1 JH817356 EKC39252.1
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000053268
UP000007266
+ More
UP000192223 UP000235965 UP000027135 UP000245037 UP000009046 UP000019118 UP000030742 UP000079169 UP000015103 UP000054359 UP000007819 UP000018468 UP000000311 UP000279307 UP000008237 UP000265200 UP000265180 UP000261540 UP000261560 UP000225706 UP000275408 UP000264820 UP000000305 UP000261620 UP000264800 UP000001038 UP000005207 UP000076858 UP000265020 UP000192220 UP000242638 UP000265040 UP000028760 UP000007635 UP000261500 UP000261640 UP000007303 UP000002852 UP000261400 UP000261420 UP000018467 UP000261440 UP000265080 UP000034805 UP000192224 UP000005226 UP000261660 UP000246464 UP000215335 UP000257200 UP000001554 UP000005408
UP000192223 UP000235965 UP000027135 UP000245037 UP000009046 UP000019118 UP000030742 UP000079169 UP000015103 UP000054359 UP000007819 UP000018468 UP000000311 UP000279307 UP000008237 UP000265200 UP000265180 UP000261540 UP000261560 UP000225706 UP000275408 UP000264820 UP000000305 UP000261620 UP000264800 UP000001038 UP000005207 UP000076858 UP000265020 UP000192220 UP000242638 UP000265040 UP000028760 UP000007635 UP000261500 UP000261640 UP000007303 UP000002852 UP000261400 UP000261420 UP000018467 UP000261440 UP000265080 UP000034805 UP000192224 UP000005226 UP000261660 UP000246464 UP000215335 UP000257200 UP000001554 UP000005408
Interpro
SUPFAM
SSF46785
SSF46785
Gene 3D
ProteinModelPortal
H9JCM2
Q2F5W8
A0A3S2TFP1
A0A2A4IZ53
A0A2H1WLC9
S4P9I7
+ More
I4DMA9 A0A194QS44 A0A194QBK9 D6WVQ2 A0A0T6B2B6 A0A1W4WEY9 A0A2J7PKP9 A0A067RIG7 A0A1B6ER76 A0A2P8XQ56 A0A1Y1MDM2 E0VNS1 N6TS51 U4TWV0 A0A0V0G3D9 A0A023F967 A0A069DZ24 A0A1S4E7U6 A0A224XP92 A0A0A9ZGG0 A0A0P4VSA9 R4G476 A0A2R7WXR0 A0A087UIR5 A0A0C5B0H1 A0A2R5LLC6 V5IA60 J9JUQ2 A0A131Z0W6 L7LZY7 A0A0C9PMI1 A0A171BB79 W5N823 E2AI04 T1J1H9 A0A131XDL0 A0A3L8DIS9 E2BW38 A0A3P9JTW7 A0A3P9LHH5 A0A1Z5KWS0 A0A131XVD4 A0A3B3RUB1 A0A0S7G0W9 A0A3B3DB81 A0A2B4SU00 A0A3M6UQ20 A0A3Q2YD84 A0A0B8RWM7 V8NWQ4 E9GVL8 A0A3Q3VQH3 A0A3Q3GI97 A0A3B3HKL0 A0A1A7WW76 I3JH38 A0A164ZVQ4 A0A3Q2GEL9 A0A2I4BWH2 A0A3P9MSX1 A0A1A8J2H2 A0A1A8DY44 A0A1A8G6P6 A0A1A8ARA8 A0A3Q1HZ04 A0A087Y5S0 A0A1A8QZC0 A0A1A8L461 A0A0N7ZJY8 A0A0P6I624 G3Q3S8 A0A3B3UV04 E9IJF3 A0A3Q3MFH1 H3CHW8 A0A0F7Z0Z0 A0A2R2YUM9 A0A3S2PH67 A0A3B5QXX0 A0A3B5AGZ2 A0A146MTG9 A0A3B4UB02 Q4T0S8 W5KWC5 A0A3B4DDI3 A0A3P8S093 A0A0P7UVL0 A0A3B5KFX6 A0A3Q3F8P0 A0A2U9BP45 A0A232FK97 A0A3Q1FXJ6 C3Z7L1 K1QQK1
I4DMA9 A0A194QS44 A0A194QBK9 D6WVQ2 A0A0T6B2B6 A0A1W4WEY9 A0A2J7PKP9 A0A067RIG7 A0A1B6ER76 A0A2P8XQ56 A0A1Y1MDM2 E0VNS1 N6TS51 U4TWV0 A0A0V0G3D9 A0A023F967 A0A069DZ24 A0A1S4E7U6 A0A224XP92 A0A0A9ZGG0 A0A0P4VSA9 R4G476 A0A2R7WXR0 A0A087UIR5 A0A0C5B0H1 A0A2R5LLC6 V5IA60 J9JUQ2 A0A131Z0W6 L7LZY7 A0A0C9PMI1 A0A171BB79 W5N823 E2AI04 T1J1H9 A0A131XDL0 A0A3L8DIS9 E2BW38 A0A3P9JTW7 A0A3P9LHH5 A0A1Z5KWS0 A0A131XVD4 A0A3B3RUB1 A0A0S7G0W9 A0A3B3DB81 A0A2B4SU00 A0A3M6UQ20 A0A3Q2YD84 A0A0B8RWM7 V8NWQ4 E9GVL8 A0A3Q3VQH3 A0A3Q3GI97 A0A3B3HKL0 A0A1A7WW76 I3JH38 A0A164ZVQ4 A0A3Q2GEL9 A0A2I4BWH2 A0A3P9MSX1 A0A1A8J2H2 A0A1A8DY44 A0A1A8G6P6 A0A1A8ARA8 A0A3Q1HZ04 A0A087Y5S0 A0A1A8QZC0 A0A1A8L461 A0A0N7ZJY8 A0A0P6I624 G3Q3S8 A0A3B3UV04 E9IJF3 A0A3Q3MFH1 H3CHW8 A0A0F7Z0Z0 A0A2R2YUM9 A0A3S2PH67 A0A3B5QXX0 A0A3B5AGZ2 A0A146MTG9 A0A3B4UB02 Q4T0S8 W5KWC5 A0A3B4DDI3 A0A3P8S093 A0A0P7UVL0 A0A3B5KFX6 A0A3Q3F8P0 A0A2U9BP45 A0A232FK97 A0A3Q1FXJ6 C3Z7L1 K1QQK1
PDB
6EPF
E-value=8.41058e-136,
Score=1240
Ontologies
KEGG
GO
PANTHER
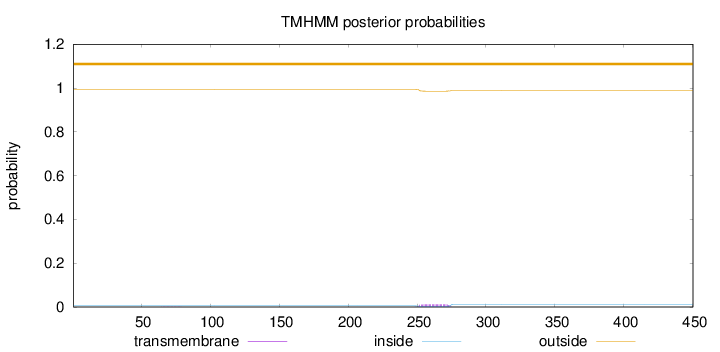
Topology
Length:
450
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.27823
Exp number, first 60 AAs:
2e-05
Total prob of N-in:
0.00560
outside
1 - 450
Population Genetic Test Statistics
Pi
222.360733
Theta
204.173353
Tajima's D
0.404046
CLR
0.069919
CSRT
0.491975401229938
Interpretation
Uncertain
