Gene
KWMTBOMO10282
Pre Gene Modal
BGIBMGA007269
Annotation
PREDICTED:_uncharacterized_protein_LOC101746561_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.778
Sequence
CDS
ATGTTAACCTTAATTATTTCTACGCTTATTTCTGCTTTAATTGTGGAAGTATTTCGTTTTTTGAATAAAAAAAGGAAGCCGGCAATATTTGGTATTTATCAACAGAAGAACAAACTTTTTTGGCCAAAATTCATATTTATGTATACAATTTTGCGCGTTAGACAATTCTTGAAGTATTTGAAACGCGAACATGCCGTTGAGGTTGGCAAAAGTGGAGACGGAAACATTCGTGTGCATGAAGAAGATAAAAAATTGGAACAGAAGTATTGTCTCGGCAGTAATCCGTTAGCAATAGACGCGGTTTATTTCAACGGAATGTCGCGAGAAGGCGATGCTGTGATTTGTGGAGTTGCTAGAAGACCTCAAAATATCTGTGATGCATTTCTATACTTGAAATTAAATAGTGAAGAGCTTCTTCTATCGCCAAATTTGCCAGATACTTGCTTGAAACAAACTGAATCAGAAGGAGGCGAATATAAAGTAAATGGAATAGAAGTTCACAATTTCATACCAATGCGAACTTGGAAACTGACTTACAATGGATCTATGAAACAAAAAAGCTATCCTGAAGATTACCTAACTGTTGAACTATCTCTAACATGGAGTGCACAATGGCCTCATTTCAATTATGACTCACATATGGCGCCATACAGTATGGCTACCGACATGGCGAGGGAATCGTGGTCGCAGGATTATTTTAAACTGCTAAAAAAGTTGCATCAAACACATTACGAACAAATGGGCGTAATTACAGGGATTGTAAAAGTTGACGGTAAACAATACGAACTGAATATGCCCGCAGTTAGAGATCATAGCTTTGGACCATTTCGCGACTGGCGCACGTTCCATCGATACGTCTACCATTTTATATTTCTTGATAACGGCGACTGTATGGCCATAGGAAGTGTTTCACAGCCGGCTATTCTGTCTCATCTCACAATTGGGTACTATTGCCGTAAGTCAGATCAAGCTGTCTTCCCTGTTGAATGGTGTGACTTCCAGCTGTACCAACACGGAGAAATGCAGACTTTGCCTAAGGATTACGGATTTATGTTCAAAGCCGGAGGTGATATTTACACGGTGAAGGTCCAAGTTGACGACGAAGACGTATTTTACATCGGAAAAGAGAGAACGTCAAAGTTTTACGAGAGATGGTCTACAGTCGATATTAATGGGGTTAAGGGACGAGCCTGTGTTGAATGGCAATATAATAATGTTTTAAATGTGACGAATAAATTGTAA
Protein
MLTLIISTLISALIVEVFRFLNKKRKPAIFGIYQQKNKLFWPKFIFMYTILRVRQFLKYLKREHAVEVGKSGDGNIRVHEEDKKLEQKYCLGSNPLAIDAVYFNGMSREGDAVICGVARRPQNICDAFLYLKLNSEELLLSPNLPDTCLKQTESEGGEYKVNGIEVHNFIPMRTWKLTYNGSMKQKSYPEDYLTVELSLTWSAQWPHFNYDSHMAPYSMATDMARESWSQDYFKLLKKLHQTHYEQMGVITGIVKVDGKQYELNMPAVRDHSFGPFRDWRTFHRYVYHFIFLDNGDCMAIGSVSQPAILSHLTIGYYCRKSDQAVFPVEWCDFQLYQHGEMQTLPKDYGFMFKAGGDIYTVKVQVDDEDVFYIGKERTSKFYERWSTVDINGVKGRACVEWQYNNVLNVTNKL
Summary
Cofactor
Zn(2+)
Similarity
Belongs to the peptidase M3 family.
Uniprot
A0A2A4J0R9
A0A2H1WN56
A0A194QCU1
A0A3S2NU99
A0A1E1WU36
A0A212EM02
+ More
A0A1B0DQG6 A0A1W4XEA8 V5H5G8 A0A0T6B096 A0A2P8ZPR9 D6X2A1 A0A1B0CAD2 A0A1B6GNV4 A0A2J7RAS1 A0A0P4YFD2 A0A0P5KQS0 A0A0P6ENC3 A0A0P6AF54 A0A0P5RUD8 A0A067QH61 A0A1S4KD98 A0A1W7R539 E9H641 A0A165AA26 A0A0P5A310 A0A1L8DAG5 A0A1L8DA56 A0A3S2NEA6 A0A0P5DNG8 Q172J6 A0A0P6I5D1 Q16Z88 A0A1B6ELM3 A0A0P5B8U6 A0A0N8D0P0 A0A0P5B489 A0A1B6KM79 A0A1B6MRU0 A0A0P5Z9I8 A0A0P5MAD7 A0A0P5J3V1 A0A0P5NQT6 A0A0N8C9L9 A0A0N1ICE3 A0A0N8E4Q7 A0A1B6M0P0 A0A0P5B5Q5 A0A0P5GSN9 A0A1E1W2I7 A0A1D2N695 A0A2A4JMP5 A0A0P6H820 D6WE14 A0A0P5HMV9 A0A0N8B1U7 A0A212FK57 A0A2W1BUJ8 A0A3R7QS53 A0A0P5ZZE6 A0A1B6E7A5 A0A0P5B4R6 A0A1W4XUH6 A0A0P5MJA2 H9JQS2 A0A0P5QIT6 A0A226ED78 A0A0P6D0T0 J3JTG1 N6T7A4 A0A0P5T1N6 A0A0P5IVP0 A0A2H1W3D2 A0A0P5LM95 A0A194Q1B1 V9KRQ7 A0A1B6LU17 A0A0P6A2B8 A0A0N7ZVM5 A0A3S0ZV74 U4UEE9 A0A2P8XU25 V4ADL6 C3Z0U3 A0A0P5TQH8 A0A2C9L2Y4 A0A0P5YS53 A0A1B6E6B9 A0A210QUV2 A0A1Y1M628 A0A267EAY8 A0A267E7Y2 R7V0T9
A0A1B0DQG6 A0A1W4XEA8 V5H5G8 A0A0T6B096 A0A2P8ZPR9 D6X2A1 A0A1B0CAD2 A0A1B6GNV4 A0A2J7RAS1 A0A0P4YFD2 A0A0P5KQS0 A0A0P6ENC3 A0A0P6AF54 A0A0P5RUD8 A0A067QH61 A0A1S4KD98 A0A1W7R539 E9H641 A0A165AA26 A0A0P5A310 A0A1L8DAG5 A0A1L8DA56 A0A3S2NEA6 A0A0P5DNG8 Q172J6 A0A0P6I5D1 Q16Z88 A0A1B6ELM3 A0A0P5B8U6 A0A0N8D0P0 A0A0P5B489 A0A1B6KM79 A0A1B6MRU0 A0A0P5Z9I8 A0A0P5MAD7 A0A0P5J3V1 A0A0P5NQT6 A0A0N8C9L9 A0A0N1ICE3 A0A0N8E4Q7 A0A1B6M0P0 A0A0P5B5Q5 A0A0P5GSN9 A0A1E1W2I7 A0A1D2N695 A0A2A4JMP5 A0A0P6H820 D6WE14 A0A0P5HMV9 A0A0N8B1U7 A0A212FK57 A0A2W1BUJ8 A0A3R7QS53 A0A0P5ZZE6 A0A1B6E7A5 A0A0P5B4R6 A0A1W4XUH6 A0A0P5MJA2 H9JQS2 A0A0P5QIT6 A0A226ED78 A0A0P6D0T0 J3JTG1 N6T7A4 A0A0P5T1N6 A0A0P5IVP0 A0A2H1W3D2 A0A0P5LM95 A0A194Q1B1 V9KRQ7 A0A1B6LU17 A0A0P6A2B8 A0A0N7ZVM5 A0A3S0ZV74 U4UEE9 A0A2P8XU25 V4ADL6 C3Z0U3 A0A0P5TQH8 A0A2C9L2Y4 A0A0P5YS53 A0A1B6E6B9 A0A210QUV2 A0A1Y1M628 A0A267EAY8 A0A267E7Y2 R7V0T9
Pubmed
EMBL
NWSH01004502
PCG65040.1
ODYU01009447
SOQ53884.1
KQ459232
KPJ02800.1
+ More
RSAL01000189 RVE44733.1 GDQN01000597 JAT90457.1 AGBW02013944 OWR42530.1 AJVK01019112 GALX01000423 JAB68043.1 LJIG01016392 KRT80738.1 PYGN01000001 PSN58506.1 KQ971371 EFA10235.2 AJWK01003658 GECZ01005668 JAS64101.1 NEVH01006565 PNF37924.1 GDIP01228381 JAI95020.1 GDIQ01182017 JAK69708.1 GDIQ01073805 JAN20932.1 GDIP01031602 JAM72113.1 GDIQ01096993 JAL54733.1 KK853736 KDQ88522.1 GEHC01001357 JAV46288.1 GL732596 EFX72741.1 LRGB01000642 KZS17357.1 GDIP01208138 JAJ15264.1 GFDF01010727 JAV03357.1 GFDF01010731 JAV03353.1 RSAL01000076 RVE48853.1 GDIP01157206 JAJ66196.1 CH477436 EAT40930.1 GDIQ01009117 JAN85620.1 CH477495 EAT39972.1 GECZ01030952 JAS38817.1 GDIP01192247 JAJ31155.1 GDIP01080327 JAM23388.1 GDIP01189891 JAJ33511.1 GEBQ01027663 JAT12314.1 GEBQ01001351 JAT38626.1 GDIP01047961 JAM55754.1 GDIQ01158367 JAK93358.1 GDIQ01205756 JAK45969.1 GDIQ01149265 JAL02461.1 GDIQ01101307 JAL50419.1 KQ460971 KPJ10127.1 GDIQ01062012 JAN32725.1 GEBQ01010486 JAT29491.1 GDIP01189890 GDIQ01126321 JAJ33512.1 JAL25405.1 GDIQ01237386 JAK14339.1 GDQN01009905 JAT81149.1 LJIJ01000186 ODN00790.1 NWSH01001090 PCG72672.1 GDIQ01035574 JAN59163.1 KQ971324 EFA01182.1 GDIQ01248098 JAK03627.1 GDIQ01221083 JAK30642.1 AGBW02008141 OWR54124.1 KZ149896 PZC78752.1 QCYY01001677 ROT76254.1 GDIP01037116 JAM66599.1 GEDC01003480 JAS33818.1 GDIP01206486 GDIP01204485 GDIQ01048603 JAJ18917.1 JAN46134.1 GDIQ01158368 JAK93357.1 BABH01012391 GDIQ01116678 JAL35048.1 LNIX01000004 OXA55370.1 GDIQ01086911 JAN07826.1 BT126517 AEE61481.1 APGK01041384 KB740993 ENN76074.1 GDIP01132299 JAL71415.1 GDIQ01207205 JAK44520.1 ODYU01006045 SOQ47547.1 GDIQ01171526 JAK80199.1 KQ459590 KPI97185.1 JW868548 AFP01066.1 GEBQ01012804 JAT27173.1 GDIP01036016 JAM67699.1 GDIP01208137 JAJ15265.1 RQTK01000131 RUS86660.1 KB632256 ERL90708.1 PYGN01001345 PSN35517.1 KB201931 ESO93220.1 GG666570 EEN53847.1 GDIP01128376 JAL75338.1 GDIP01055315 JAM48400.1 GEDC01003843 JAS33455.1 NEDP02001786 OWF52519.1 GEZM01041364 JAV80478.1 NIVC01002439 PAA57872.1 NIVC01002571 PAA56802.1 AMQN01006444 KB298361 ELU09306.1
RSAL01000189 RVE44733.1 GDQN01000597 JAT90457.1 AGBW02013944 OWR42530.1 AJVK01019112 GALX01000423 JAB68043.1 LJIG01016392 KRT80738.1 PYGN01000001 PSN58506.1 KQ971371 EFA10235.2 AJWK01003658 GECZ01005668 JAS64101.1 NEVH01006565 PNF37924.1 GDIP01228381 JAI95020.1 GDIQ01182017 JAK69708.1 GDIQ01073805 JAN20932.1 GDIP01031602 JAM72113.1 GDIQ01096993 JAL54733.1 KK853736 KDQ88522.1 GEHC01001357 JAV46288.1 GL732596 EFX72741.1 LRGB01000642 KZS17357.1 GDIP01208138 JAJ15264.1 GFDF01010727 JAV03357.1 GFDF01010731 JAV03353.1 RSAL01000076 RVE48853.1 GDIP01157206 JAJ66196.1 CH477436 EAT40930.1 GDIQ01009117 JAN85620.1 CH477495 EAT39972.1 GECZ01030952 JAS38817.1 GDIP01192247 JAJ31155.1 GDIP01080327 JAM23388.1 GDIP01189891 JAJ33511.1 GEBQ01027663 JAT12314.1 GEBQ01001351 JAT38626.1 GDIP01047961 JAM55754.1 GDIQ01158367 JAK93358.1 GDIQ01205756 JAK45969.1 GDIQ01149265 JAL02461.1 GDIQ01101307 JAL50419.1 KQ460971 KPJ10127.1 GDIQ01062012 JAN32725.1 GEBQ01010486 JAT29491.1 GDIP01189890 GDIQ01126321 JAJ33512.1 JAL25405.1 GDIQ01237386 JAK14339.1 GDQN01009905 JAT81149.1 LJIJ01000186 ODN00790.1 NWSH01001090 PCG72672.1 GDIQ01035574 JAN59163.1 KQ971324 EFA01182.1 GDIQ01248098 JAK03627.1 GDIQ01221083 JAK30642.1 AGBW02008141 OWR54124.1 KZ149896 PZC78752.1 QCYY01001677 ROT76254.1 GDIP01037116 JAM66599.1 GEDC01003480 JAS33818.1 GDIP01206486 GDIP01204485 GDIQ01048603 JAJ18917.1 JAN46134.1 GDIQ01158368 JAK93357.1 BABH01012391 GDIQ01116678 JAL35048.1 LNIX01000004 OXA55370.1 GDIQ01086911 JAN07826.1 BT126517 AEE61481.1 APGK01041384 KB740993 ENN76074.1 GDIP01132299 JAL71415.1 GDIQ01207205 JAK44520.1 ODYU01006045 SOQ47547.1 GDIQ01171526 JAK80199.1 KQ459590 KPI97185.1 JW868548 AFP01066.1 GEBQ01012804 JAT27173.1 GDIP01036016 JAM67699.1 GDIP01208137 JAJ15265.1 RQTK01000131 RUS86660.1 KB632256 ERL90708.1 PYGN01001345 PSN35517.1 KB201931 ESO93220.1 GG666570 EEN53847.1 GDIP01128376 JAL75338.1 GDIP01055315 JAM48400.1 GEDC01003843 JAS33455.1 NEDP02001786 OWF52519.1 GEZM01041364 JAV80478.1 NIVC01002439 PAA57872.1 NIVC01002571 PAA56802.1 AMQN01006444 KB298361 ELU09306.1
Proteomes
UP000218220
UP000053268
UP000283053
UP000007151
UP000092462
UP000192223
+ More
UP000245037 UP000007266 UP000092461 UP000235965 UP000027135 UP000000305 UP000076858 UP000008820 UP000053240 UP000094527 UP000283509 UP000005204 UP000198287 UP000019118 UP000271974 UP000030742 UP000030746 UP000001554 UP000076420 UP000242188 UP000215902 UP000014760
UP000245037 UP000007266 UP000092461 UP000235965 UP000027135 UP000000305 UP000076858 UP000008820 UP000053240 UP000094527 UP000283509 UP000005204 UP000198287 UP000019118 UP000271974 UP000030742 UP000030746 UP000001554 UP000076420 UP000242188 UP000215902 UP000014760
PRIDE
Interpro
SUPFAM
SSF103473
SSF103473
Gene 3D
CDD
ProteinModelPortal
A0A2A4J0R9
A0A2H1WN56
A0A194QCU1
A0A3S2NU99
A0A1E1WU36
A0A212EM02
+ More
A0A1B0DQG6 A0A1W4XEA8 V5H5G8 A0A0T6B096 A0A2P8ZPR9 D6X2A1 A0A1B0CAD2 A0A1B6GNV4 A0A2J7RAS1 A0A0P4YFD2 A0A0P5KQS0 A0A0P6ENC3 A0A0P6AF54 A0A0P5RUD8 A0A067QH61 A0A1S4KD98 A0A1W7R539 E9H641 A0A165AA26 A0A0P5A310 A0A1L8DAG5 A0A1L8DA56 A0A3S2NEA6 A0A0P5DNG8 Q172J6 A0A0P6I5D1 Q16Z88 A0A1B6ELM3 A0A0P5B8U6 A0A0N8D0P0 A0A0P5B489 A0A1B6KM79 A0A1B6MRU0 A0A0P5Z9I8 A0A0P5MAD7 A0A0P5J3V1 A0A0P5NQT6 A0A0N8C9L9 A0A0N1ICE3 A0A0N8E4Q7 A0A1B6M0P0 A0A0P5B5Q5 A0A0P5GSN9 A0A1E1W2I7 A0A1D2N695 A0A2A4JMP5 A0A0P6H820 D6WE14 A0A0P5HMV9 A0A0N8B1U7 A0A212FK57 A0A2W1BUJ8 A0A3R7QS53 A0A0P5ZZE6 A0A1B6E7A5 A0A0P5B4R6 A0A1W4XUH6 A0A0P5MJA2 H9JQS2 A0A0P5QIT6 A0A226ED78 A0A0P6D0T0 J3JTG1 N6T7A4 A0A0P5T1N6 A0A0P5IVP0 A0A2H1W3D2 A0A0P5LM95 A0A194Q1B1 V9KRQ7 A0A1B6LU17 A0A0P6A2B8 A0A0N7ZVM5 A0A3S0ZV74 U4UEE9 A0A2P8XU25 V4ADL6 C3Z0U3 A0A0P5TQH8 A0A2C9L2Y4 A0A0P5YS53 A0A1B6E6B9 A0A210QUV2 A0A1Y1M628 A0A267EAY8 A0A267E7Y2 R7V0T9
A0A1B0DQG6 A0A1W4XEA8 V5H5G8 A0A0T6B096 A0A2P8ZPR9 D6X2A1 A0A1B0CAD2 A0A1B6GNV4 A0A2J7RAS1 A0A0P4YFD2 A0A0P5KQS0 A0A0P6ENC3 A0A0P6AF54 A0A0P5RUD8 A0A067QH61 A0A1S4KD98 A0A1W7R539 E9H641 A0A165AA26 A0A0P5A310 A0A1L8DAG5 A0A1L8DA56 A0A3S2NEA6 A0A0P5DNG8 Q172J6 A0A0P6I5D1 Q16Z88 A0A1B6ELM3 A0A0P5B8U6 A0A0N8D0P0 A0A0P5B489 A0A1B6KM79 A0A1B6MRU0 A0A0P5Z9I8 A0A0P5MAD7 A0A0P5J3V1 A0A0P5NQT6 A0A0N8C9L9 A0A0N1ICE3 A0A0N8E4Q7 A0A1B6M0P0 A0A0P5B5Q5 A0A0P5GSN9 A0A1E1W2I7 A0A1D2N695 A0A2A4JMP5 A0A0P6H820 D6WE14 A0A0P5HMV9 A0A0N8B1U7 A0A212FK57 A0A2W1BUJ8 A0A3R7QS53 A0A0P5ZZE6 A0A1B6E7A5 A0A0P5B4R6 A0A1W4XUH6 A0A0P5MJA2 H9JQS2 A0A0P5QIT6 A0A226ED78 A0A0P6D0T0 J3JTG1 N6T7A4 A0A0P5T1N6 A0A0P5IVP0 A0A2H1W3D2 A0A0P5LM95 A0A194Q1B1 V9KRQ7 A0A1B6LU17 A0A0P6A2B8 A0A0N7ZVM5 A0A3S0ZV74 U4UEE9 A0A2P8XU25 V4ADL6 C3Z0U3 A0A0P5TQH8 A0A2C9L2Y4 A0A0P5YS53 A0A1B6E6B9 A0A210QUV2 A0A1Y1M628 A0A267EAY8 A0A267E7Y2 R7V0T9
Ontologies
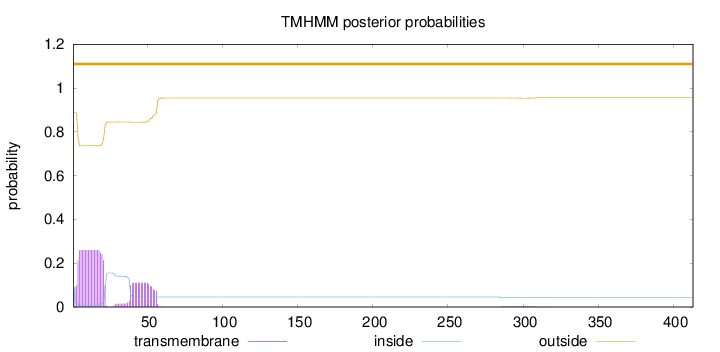
Topology
Length:
413
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.94201
Exp number, first 60 AAs:
6.85851
Total prob of N-in:
0.11299
outside
1 - 413
Population Genetic Test Statistics
Pi
238.639163
Theta
178.763111
Tajima's D
1.401424
CLR
0.460101
CSRT
0.763161841907905
Interpretation
Uncertain
