Gene
KWMTBOMO10280
Pre Gene Modal
BGIBMGA007291
Annotation
PREDICTED:_uncharacterized_protein_LOC106107222_[Papilio_polytes]
Location in the cell
PlasmaMembrane Reliability : 3.3
Sequence
CDS
ATGGATTTACGCATTGCTGTATTAGTTTTATGTGCACTTTTTGGGAAAGCCGTCGGTCGATTCAGATGCTATGCATGTAGTTTTTCATCAGTGGATTCGGATCAATCGTGCCTGACAATAGGAAACAACACGAACATCGTGGAGTGTCCATTCACTTATTGCACAATTACGAGACAAGAGTTCATAGACCCAATAGGCGTGATCGCAAGCTTTACACGGGGCTGCGAAAGTTTCCCGGATTTTTTAAATCACGAAGTTACAGATCCAACATTCCGAACTTTTTATAGAGCATGCACAAATGATTTGTGCAACATTGGTGATGGAATTCAGTCTATAGTCGGCGGTAATTTGTCTCCGTATCCTGCATACACCGGCATTAATCTATTAGTACCTGGGACAGGAAGCGGAGTAAATATCAAAATGAATAGAGTGTTGTTCTTTTCTGTGATAATGATCCTAATCGGGTTACATGTTTAA
Protein
MDLRIAVLVLCALFGKAVGRFRCYACSFSSVDSDQSCLTIGNNTNIVECPFTYCTITRQEFIDPIGVIASFTRGCESFPDFLNHEVTDPTFRTFYRACTNDLCNIGDGIQSIVGGNLSPYPAYTGINLLVPGTGSGVNIKMNRVLFFSVIMILIGLHV
Summary
Uniprot
H9JCP5
A0A194QH81
A0A2A4IYL9
A0A2H1WLC4
A0A3S2M0T9
A0A212EM29
+ More
A0A1J1J3C8 A0A023EH46 A0A182H2E9 A0A182INH6 A0A182K022 A0A182XZE5 A0A182P9Y5 A0A182WIW4 A0A084VF25 A0A182UWT6 A0A182KRZ1 A0A182X6J0 Q5TX43 W5JU33 A0A0K8VA09 A0A034W7H8 A0A182I3T9 A0A182FFX9 A0A1S4FTM6 A0A139WKZ1 A0A182NHE7 Q16PM2 A0A0A1XHR5 A0A336LPS4 A0A1L8DBH1 A0A1L8DNV6 A0A1A9YEW2 A0A1B0CYJ9 A8CAC7 A0A1B0GAA1 A0A1B0BKG0 D3TLK9 A0A1A9UZC9 A0A1A9ZGV7 A0A0C9PJ93 A0A1B0CCM4 N6TNX0 A0A182RB39 A0A182TMQ6 E9FTZ4 A0A3B0JK67 A0A0P6JTR4 B4LLV3 A0A0P5T9F7 A0A164LNS5 A0A1Y1JYB9 A0A0P6AZH3 A0A0J9RGL1 E9FTZ3 B4QDP5 B4PBM7 B3MGI1 B4MJJ0 B4HPE6 A1ZBJ7 A0A0N8DE84 B3NJI7 D5SHM1 A0A226DEF7 A0A0N8D1S9 R7U1I8 A0A0P6DWB9 A0A0P5Y4R7 A0A3R7QCM5 D6WWJ8 A0A0P5TG38
A0A1J1J3C8 A0A023EH46 A0A182H2E9 A0A182INH6 A0A182K022 A0A182XZE5 A0A182P9Y5 A0A182WIW4 A0A084VF25 A0A182UWT6 A0A182KRZ1 A0A182X6J0 Q5TX43 W5JU33 A0A0K8VA09 A0A034W7H8 A0A182I3T9 A0A182FFX9 A0A1S4FTM6 A0A139WKZ1 A0A182NHE7 Q16PM2 A0A0A1XHR5 A0A336LPS4 A0A1L8DBH1 A0A1L8DNV6 A0A1A9YEW2 A0A1B0CYJ9 A8CAC7 A0A1B0GAA1 A0A1B0BKG0 D3TLK9 A0A1A9UZC9 A0A1A9ZGV7 A0A0C9PJ93 A0A1B0CCM4 N6TNX0 A0A182RB39 A0A182TMQ6 E9FTZ4 A0A3B0JK67 A0A0P6JTR4 B4LLV3 A0A0P5T9F7 A0A164LNS5 A0A1Y1JYB9 A0A0P6AZH3 A0A0J9RGL1 E9FTZ3 B4QDP5 B4PBM7 B3MGI1 B4MJJ0 B4HPE6 A1ZBJ7 A0A0N8DE84 B3NJI7 D5SHM1 A0A226DEF7 A0A0N8D1S9 R7U1I8 A0A0P6DWB9 A0A0P5Y4R7 A0A3R7QCM5 D6WWJ8 A0A0P5TG38
Pubmed
19121390
26354079
22118469
24945155
26483478
25244985
+ More
24438588 20966253 12364791 14747013 17210077 20920257 23761445 25348373 18362917 19820115 17510324 25830018 17760985 20353571 23537049 21292972 17994087 28004739 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 23254933
24438588 20966253 12364791 14747013 17210077 20920257 23761445 25348373 18362917 19820115 17510324 25830018 17760985 20353571 23537049 21292972 17994087 28004739 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 23254933
EMBL
BABH01034651
BABH01034652
BABH01034653
KQ459232
KPJ02801.1
NWSH01004502
+ More
PCG65037.1 ODYU01009447 SOQ53883.1 RSAL01000083 RVE48466.1 AGBW02013944 OWR42529.1 CVRI01000067 CRL06864.1 GAPW01005417 JAC08181.1 JXUM01105243 KQ564950 KXJ71517.1 ATLV01012360 KE524785 KFB36569.1 AAAB01008807 EAL41723.1 ADMH02000483 ETN66274.1 GDHF01016919 JAI35395.1 GAKP01009254 JAC49698.1 APCN01000221 KQ971323 KYB28554.1 CH477777 EAT36317.1 GBXI01004244 JAD10048.1 UFQT01000100 SSX19940.1 GFDF01010276 JAV03808.1 GFDF01005942 JAV08142.1 AJVK01001940 EU040042 ABV44732.1 CCAG010018535 JXJN01015943 EZ422311 ADD18587.1 GBYB01001033 JAG70800.1 AJWK01006917 AJWK01006918 APGK01057468 APGK01057469 KB741280 ENN70935.1 GL732524 EFX89554.1 OUUW01000001 SPP73011.1 GDIQ01008743 JAN85994.1 CH940648 EDW61976.2 GDIP01130760 JAL72954.1 LRGB01003123 KZS04293.1 GEZM01100791 JAV52790.1 GDIP01022232 JAM81483.1 CM002911 KMY95071.1 EFX89555.1 CM000362 EDX07801.1 CM000158 EDW91511.2 CH902619 EDV35724.2 CH963846 EDW72279.2 CH480816 EDW48584.1 AE013599 ABC66049.2 GDIP01042375 JAM61340.1 CH954179 EDV55195.2 BT124822 ADF87917.1 LNIX01000021 OXA43549.1 GDIP01077215 JAM26500.1 AMQN01001868 KB306459 ELT99814.1 GDIQ01075171 JAN19566.1 GDIP01063970 JAM39745.1 QCYY01001878 ROT74552.1 KQ971361 EFA08720.1 GDIP01126714 JAL77000.1
PCG65037.1 ODYU01009447 SOQ53883.1 RSAL01000083 RVE48466.1 AGBW02013944 OWR42529.1 CVRI01000067 CRL06864.1 GAPW01005417 JAC08181.1 JXUM01105243 KQ564950 KXJ71517.1 ATLV01012360 KE524785 KFB36569.1 AAAB01008807 EAL41723.1 ADMH02000483 ETN66274.1 GDHF01016919 JAI35395.1 GAKP01009254 JAC49698.1 APCN01000221 KQ971323 KYB28554.1 CH477777 EAT36317.1 GBXI01004244 JAD10048.1 UFQT01000100 SSX19940.1 GFDF01010276 JAV03808.1 GFDF01005942 JAV08142.1 AJVK01001940 EU040042 ABV44732.1 CCAG010018535 JXJN01015943 EZ422311 ADD18587.1 GBYB01001033 JAG70800.1 AJWK01006917 AJWK01006918 APGK01057468 APGK01057469 KB741280 ENN70935.1 GL732524 EFX89554.1 OUUW01000001 SPP73011.1 GDIQ01008743 JAN85994.1 CH940648 EDW61976.2 GDIP01130760 JAL72954.1 LRGB01003123 KZS04293.1 GEZM01100791 JAV52790.1 GDIP01022232 JAM81483.1 CM002911 KMY95071.1 EFX89555.1 CM000362 EDX07801.1 CM000158 EDW91511.2 CH902619 EDV35724.2 CH963846 EDW72279.2 CH480816 EDW48584.1 AE013599 ABC66049.2 GDIP01042375 JAM61340.1 CH954179 EDV55195.2 BT124822 ADF87917.1 LNIX01000021 OXA43549.1 GDIP01077215 JAM26500.1 AMQN01001868 KB306459 ELT99814.1 GDIQ01075171 JAN19566.1 GDIP01063970 JAM39745.1 QCYY01001878 ROT74552.1 KQ971361 EFA08720.1 GDIP01126714 JAL77000.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000283053
UP000007151
UP000183832
+ More
UP000069940 UP000249989 UP000075880 UP000075881 UP000076408 UP000075885 UP000075920 UP000030765 UP000075903 UP000075882 UP000076407 UP000007062 UP000000673 UP000075840 UP000069272 UP000007266 UP000075884 UP000008820 UP000092443 UP000092462 UP000092444 UP000092460 UP000078200 UP000092445 UP000092461 UP000019118 UP000075900 UP000075902 UP000000305 UP000268350 UP000008792 UP000076858 UP000000304 UP000002282 UP000007801 UP000007798 UP000001292 UP000000803 UP000008711 UP000198287 UP000014760 UP000283509
UP000069940 UP000249989 UP000075880 UP000075881 UP000076408 UP000075885 UP000075920 UP000030765 UP000075903 UP000075882 UP000076407 UP000007062 UP000000673 UP000075840 UP000069272 UP000007266 UP000075884 UP000008820 UP000092443 UP000092462 UP000092444 UP000092460 UP000078200 UP000092445 UP000092461 UP000019118 UP000075900 UP000075902 UP000000305 UP000268350 UP000008792 UP000076858 UP000000304 UP000002282 UP000007801 UP000007798 UP000001292 UP000000803 UP000008711 UP000198287 UP000014760 UP000283509
Pfam
PF03372 Exo_endo_phos
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
H9JCP5
A0A194QH81
A0A2A4IYL9
A0A2H1WLC4
A0A3S2M0T9
A0A212EM29
+ More
A0A1J1J3C8 A0A023EH46 A0A182H2E9 A0A182INH6 A0A182K022 A0A182XZE5 A0A182P9Y5 A0A182WIW4 A0A084VF25 A0A182UWT6 A0A182KRZ1 A0A182X6J0 Q5TX43 W5JU33 A0A0K8VA09 A0A034W7H8 A0A182I3T9 A0A182FFX9 A0A1S4FTM6 A0A139WKZ1 A0A182NHE7 Q16PM2 A0A0A1XHR5 A0A336LPS4 A0A1L8DBH1 A0A1L8DNV6 A0A1A9YEW2 A0A1B0CYJ9 A8CAC7 A0A1B0GAA1 A0A1B0BKG0 D3TLK9 A0A1A9UZC9 A0A1A9ZGV7 A0A0C9PJ93 A0A1B0CCM4 N6TNX0 A0A182RB39 A0A182TMQ6 E9FTZ4 A0A3B0JK67 A0A0P6JTR4 B4LLV3 A0A0P5T9F7 A0A164LNS5 A0A1Y1JYB9 A0A0P6AZH3 A0A0J9RGL1 E9FTZ3 B4QDP5 B4PBM7 B3MGI1 B4MJJ0 B4HPE6 A1ZBJ7 A0A0N8DE84 B3NJI7 D5SHM1 A0A226DEF7 A0A0N8D1S9 R7U1I8 A0A0P6DWB9 A0A0P5Y4R7 A0A3R7QCM5 D6WWJ8 A0A0P5TG38
A0A1J1J3C8 A0A023EH46 A0A182H2E9 A0A182INH6 A0A182K022 A0A182XZE5 A0A182P9Y5 A0A182WIW4 A0A084VF25 A0A182UWT6 A0A182KRZ1 A0A182X6J0 Q5TX43 W5JU33 A0A0K8VA09 A0A034W7H8 A0A182I3T9 A0A182FFX9 A0A1S4FTM6 A0A139WKZ1 A0A182NHE7 Q16PM2 A0A0A1XHR5 A0A336LPS4 A0A1L8DBH1 A0A1L8DNV6 A0A1A9YEW2 A0A1B0CYJ9 A8CAC7 A0A1B0GAA1 A0A1B0BKG0 D3TLK9 A0A1A9UZC9 A0A1A9ZGV7 A0A0C9PJ93 A0A1B0CCM4 N6TNX0 A0A182RB39 A0A182TMQ6 E9FTZ4 A0A3B0JK67 A0A0P6JTR4 B4LLV3 A0A0P5T9F7 A0A164LNS5 A0A1Y1JYB9 A0A0P6AZH3 A0A0J9RGL1 E9FTZ3 B4QDP5 B4PBM7 B3MGI1 B4MJJ0 B4HPE6 A1ZBJ7 A0A0N8DE84 B3NJI7 D5SHM1 A0A226DEF7 A0A0N8D1S9 R7U1I8 A0A0P6DWB9 A0A0P5Y4R7 A0A3R7QCM5 D6WWJ8 A0A0P5TG38
Ontologies
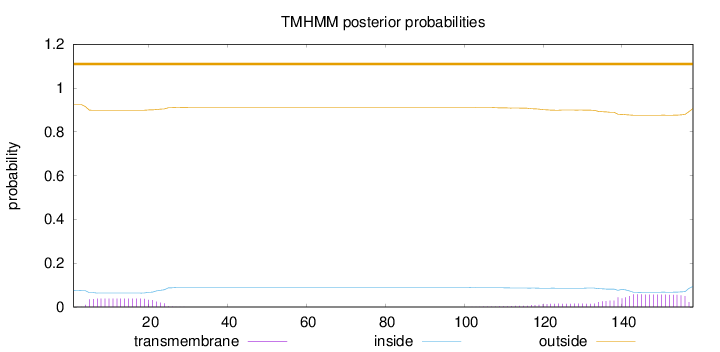
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.992896
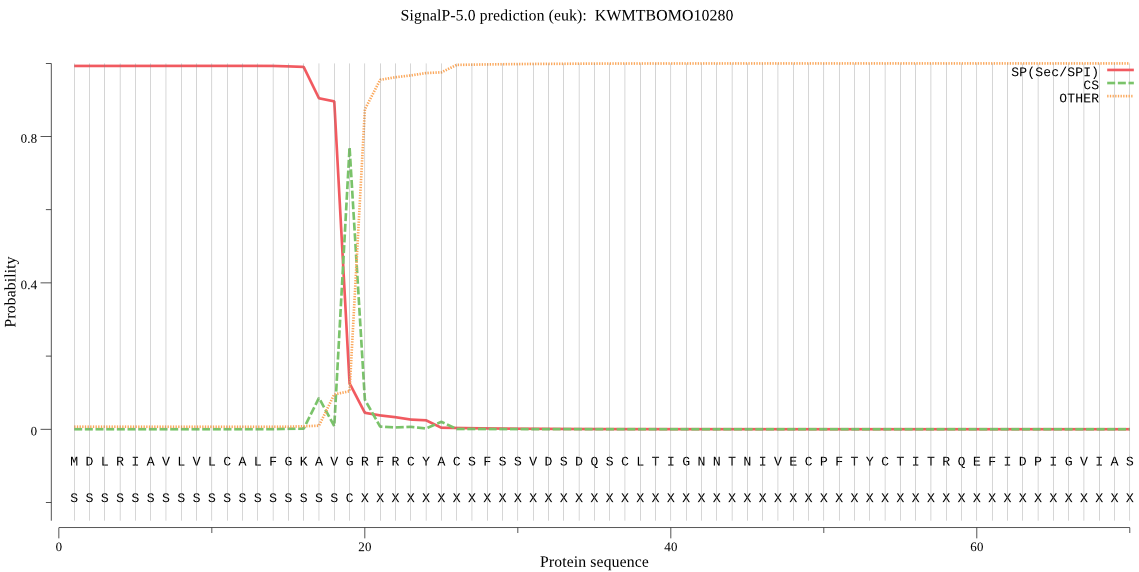
Length:
158
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.10033
Exp number, first 60 AAs:
0.707
Total prob of N-in:
0.07571
outside
1 - 158
Population Genetic Test Statistics
Pi
203.127722
Theta
172.732134
Tajima's D
0.495922
CLR
0
CSRT
0.510174491275436
Interpretation
Uncertain
