Gene
KWMTBOMO10278
Pre Gene Modal
BGIBMGA007290
Annotation
PREDICTED:_uncharacterized_protein_LOC101739820?_partial_[Bombyx_mori]
Full name
Chloride channel protein
Location in the cell
PlasmaMembrane Reliability : 4.052
Sequence
CDS
ATGCTACAGAGAAATAGTAGTGAAGGTATTTTTATACAATCAATGGATGATATAATGGATTATGATAAGTTAAAATTTGAGCATCAAGAACATCAGATTTTATTATTGGTGGATTTGAGATGCCTGAATCAGAGTAAGAATATGGCAAAGATAACTGATTATCTACAATCCGGTGCACCGTTTCGATGGATCTTTATTAGTAACACTGAAATCGAAAACCTCATTCCACCAGAAAACACAATAATGAGAATGAACATTTTACCGAATACCGAAGCTTTATTTATTCAAAGAACAGATAATAGATGCTACGAAATCAGTATGGTTTACAAAATCGGAACCGAGACTGCATGGAAAACAGAAAAGTACGCTTATTGGAATATGAGTGGAGGTTTTCAGAAAAATACCAACATTAATGAAGTTATAGCTGTAAGGAGACGTGATTTGGAAGGATACGAGATAAAAATTTGTTACGTTTTGACGGACAACGATAGTATTCATCATTTGAGTGATGAAGTGAATGATCATATTGATACGATAACTAAGGTCAATTTCCCAAGCACCAACCATCTGTTAGACTTTCTGAATGCTGAACGAAAATATGTTTTTGTTAATACGTGGGGCTATCGAATTAACGGCACTTGGAATGGACTAACTGGATTTTTGGTAAACGGCGATGTTGAAATTGGAGGGTCACCGATGTTTTTCACTGCAGAACGTACAGCGGTCGTAGATTTCATTTCGAGCCCTACACCAACTCGCTCGAAATTTGTATTTCAACAACCAAAGTTGTCGTATGAAAATAATTTATTTTTACTTTCATTTCGTACAGCCGTCTGGTATAGCACTCTAGCGTTAATCTCCTTAATATTCACAATGTTGCTGGCCGTTACAGCTTGGGAATGGAAGAAGATGTCCCAAATCAAAACGAGAGACATCGATGCTGGCGTACTTAGGCCGAGCGTTACAGATGTTACCATGCTAGTTTTTGGTGCTACTTGCCAACAAGGAAGTACCGTTGAATTAAAAGGTTCGCTCGGTCGTGTCGTAATGTTAATTCTTTTCCTGACCCTTATGTTCCTGTACACTTCATATTCAGCAAATATCGTTGCTTTACTTCAGTCCAGTTCATCCCAAATTAAAACACTGGAAGACCTTTTGCATTCCCGATTGAAATTCGGTGTACACGACACAGTTTTCAATAGATATTACTTTTCAACTGCAGATGAACCTGTTAGGAAAGCAATATATGAAAAGAAAATTGCACCCCCAGGTGTTGCACCTCAGTTTATGTCTATGGAAGAAGGTGTTAAGAAGATGCGTAAGGGTCTGTTCGCATTCCATATGGAAACAGGAGTTGGATATAAATTCGTTGGAAAATATTTCAAAGAAAGCGAAAAATGTGGCTTAAAGGAAATACAATACTTACAAGTCATAGATCCGTGGTTAGCTGTCCGCAAGAACACACCGTACAAGGAAATGTTCAAAATTGGGTAA
Protein
MLQRNSSEGIFIQSMDDIMDYDKLKFEHQEHQILLLVDLRCLNQSKNMAKITDYLQSGAPFRWIFISNTEIENLIPPENTIMRMNILPNTEALFIQRTDNRCYEISMVYKIGTETAWKTEKYAYWNMSGGFQKNTNINEVIAVRRRDLEGYEIKICYVLTDNDSIHHLSDEVNDHIDTITKVNFPSTNHLLDFLNAERKYVFVNTWGYRINGTWNGLTGFLVNGDVEIGGSPMFFTAERTAVVDFISSPTPTRSKFVFQQPKLSYENNLFLLSFRTAVWYSTLALISLIFTMLLAVTAWEWKKMSQIKTRDIDAGVLRPSVTDVTMLVFGATCQQGSTVELKGSLGRVVMLILFLTLMFLYTSYSANIVALLQSSSSQIKTLEDLLHSRLKFGVHDTVFNRYYFSTADEPVRKAIYEKKIAPPGVAPQFMSMEEGVKKMRKGLFAFHMETGVGYKFVGKYFKESEKCGLKEIQYLQVIDPWLAVRKNTPYKEMFKIG
Summary
Similarity
Belongs to the chloride channel (TC 2.A.49) family.
Uniprot
A0A386H9K5
A0A1W5LAS2
A0A0F7QIG2
A0A2D0WKS5
A0A0S1TQ33
A0A223HCX7
+ More
A0A3S2L3E3 A0A1B3B728 A0A146JYY8 A0A1Q1PP98 A0A2H1VST4 H9A5R2 A0A345BF14 A0A0K8TUD4 A0A223HDB2 E5FIB0 A0A212FEZ5 A0A140G9H5 A0A2A4IYY8 A0A2K8GL75 A0A076E9E7 A0A194QBG6 A0A194QT07 A0A0U3C6J1 A0A0S3J2Z8 A0A1B3B7C4 A0A182XCS1 A0A182RN63 A0A182PN91 A0A182HM47 A0A223HCY5 A0A0F7QIG0 A0A345BF17 A0A2A4IZW1 A0A182NM91 A0A223HDB5 A0A182M3R5 A0A1Q1PP97 A0A182Y0V0 A0A139WJQ1 A0A182GL17 Q178H7 A0A182TLZ3 A0A1S4FBU2 H9A5R6 F5HLW5 A0A1J0KKU9 Q7Q771 A0A1J1IJ30 A0A1W6L166 D6X2Y3 A0A182FFC7 A0A0C5DMP4 U4UF86 N6U3W0 A0A1V1WBQ2 A0A182KER7 A0A2D0WKS7 A0A2K8GL72 A0A182J1B5 A0A2P9JY95 A0A084W2V2 A0A2H1VRQ6 A0A1J0KKK8 A0A310SDV6 A0A1B1FKH3 A0A146JVT1 A0A0E3Y709 A0A2H4ZBE9 A0A0K8TUC6 A0A067R2S3 A0A140G9H6 A0A0U5APN7 A0A067QZN3 A0A2P8YXS4 Q16HB2 A0A2P8YXT7 A0A1B3P5F9 A0A2P8YXT1 A0A2P8XXS9 A0A182GF22 A0A182QH81 A0A232FEK6 A0A140G9H3 A0A1J0KL22 A0A1Q1PP74 A0A182FF56 A0A2J7QJP5 A0A2J7QJP1
A0A3S2L3E3 A0A1B3B728 A0A146JYY8 A0A1Q1PP98 A0A2H1VST4 H9A5R2 A0A345BF14 A0A0K8TUD4 A0A223HDB2 E5FIB0 A0A212FEZ5 A0A140G9H5 A0A2A4IYY8 A0A2K8GL75 A0A076E9E7 A0A194QBG6 A0A194QT07 A0A0U3C6J1 A0A0S3J2Z8 A0A1B3B7C4 A0A182XCS1 A0A182RN63 A0A182PN91 A0A182HM47 A0A223HCY5 A0A0F7QIG0 A0A345BF17 A0A2A4IZW1 A0A182NM91 A0A223HDB5 A0A182M3R5 A0A1Q1PP97 A0A182Y0V0 A0A139WJQ1 A0A182GL17 Q178H7 A0A182TLZ3 A0A1S4FBU2 H9A5R6 F5HLW5 A0A1J0KKU9 Q7Q771 A0A1J1IJ30 A0A1W6L166 D6X2Y3 A0A182FFC7 A0A0C5DMP4 U4UF86 N6U3W0 A0A1V1WBQ2 A0A182KER7 A0A2D0WKS7 A0A2K8GL72 A0A182J1B5 A0A2P9JY95 A0A084W2V2 A0A2H1VRQ6 A0A1J0KKK8 A0A310SDV6 A0A1B1FKH3 A0A146JVT1 A0A0E3Y709 A0A2H4ZBE9 A0A0K8TUC6 A0A067R2S3 A0A140G9H6 A0A0U5APN7 A0A067QZN3 A0A2P8YXS4 Q16HB2 A0A2P8YXT7 A0A1B3P5F9 A0A2P8YXT1 A0A2P8XXS9 A0A182GF22 A0A182QH81 A0A232FEK6 A0A140G9H3 A0A1J0KL22 A0A1Q1PP74 A0A182FF56 A0A2J7QJP5 A0A2J7QJP1
Pubmed
25803580
26812239
28150741
22363688
27006164
29727827
+ More
26017144 21091811 22118469 27004525 29375398 24998398 26354079 26657286 26626891 27171401 25244985 18362917 19820115 26483478 17510324 12364791 27836740 25665775 23537049 24438588 25856077 29133804 24845553 26760975 29403074 27538507 28648823
26017144 21091811 22118469 27004525 29375398 24998398 26354079 26657286 26626891 27171401 25244985 18362917 19820115 26483478 17510324 12364791 27836740 25665775 23537049 24438588 25856077 29133804 24845553 26760975 29403074 27538507 28648823
EMBL
MG546648
AYD42271.1
KT991368
ANA75034.2
LC017794
BAR64808.1
+ More
KY019179 ARB05668.1 KR912013 ALM24940.1 KY283576 AST36236.1 RSAL01000189 RVE44730.1 KT588085 AOE47995.1 GEDO01000005 JAP88621.1 KY325456 AQM73617.1 ODYU01004051 SOQ43532.1 JN836712 AFC91752.1 MG820667 AXF48838.1 GCVX01000138 JAI18092.1 KY283705 AST36364.1 HM562974 ADR64685.1 AGBW02008878 OWR52312.1 KU702627 AMM70654.1 NWSH01004502 PCG65035.1 KY225535 ARO70547.1 KF487731 AII01129.1 KQ459232 KPJ02802.1 KQ461155 KPJ08135.1 KP975110 ALT31629.1 KT381535 ALR72541.1 KU291857 AOE48107.1 APCN01004347 KY283575 AST36235.1 LC017789 BAR64803.1 MG820670 AXF48841.1 PCG65036.1 KY283704 AST36363.1 AXCM01010715 KY325455 AQM73616.1 KQ971338 KYB28047.1 JXUM01071488 KQ562666 KXJ75363.1 CH477363 EAT42606.1 JN836716 AFC91756.2 AAAB01008960 EGK97276.1 KX298830 APC94347.1 EAA11187.5 CVRI01000054 CRL00245.1 KX609444 KB465935 ARN17851.1 RLZ02208.1 KQ971372 EFA10663.2 KP296776 AJO62240.1 KB632161 ERL89256.1 APGK01043551 APGK01043552 APGK01043553 KB741015 ENN75306.1 GENK01000124 JAV45789.1 KY019178 ARB05667.1 KY225540 ARO70552.1 KY817085 AVH87301.1 ATLV01019703 KE525278 KFB44546.1 SOQ43533.1 KX290685 APC94256.1 GGOB01000048 MCH29444.1 KT268301 ANQ46495.1 GEDO01000004 JAP88622.1 KM251708 AKC58589.1 MF975509 AUF73085.1 GCVX01000136 JAI18094.1 KK852952 KDR13328.1 KU702628 AMM70655.1 FX982942 BAU20268.1 KK852854 KDR14986.1 PYGN01000296 PSN49053.1 CH478187 EAT33629.1 PSN49052.1 KX655899 AOG12848.1 PSN49051.1 PYGN01001184 PSN36819.1 JXUM01059078 JXUM01059079 JXUM01059080 JXUM01059081 JXUM01059082 KQ562034 KXJ76850.1 AXCN02000940 NNAY01000335 OXU29125.1 KU702625 AMM70652.1 KX298831 APC94348.1 KY325454 AQM73615.1 NEVH01013550 PNF28810.1 PNF28811.1
KY019179 ARB05668.1 KR912013 ALM24940.1 KY283576 AST36236.1 RSAL01000189 RVE44730.1 KT588085 AOE47995.1 GEDO01000005 JAP88621.1 KY325456 AQM73617.1 ODYU01004051 SOQ43532.1 JN836712 AFC91752.1 MG820667 AXF48838.1 GCVX01000138 JAI18092.1 KY283705 AST36364.1 HM562974 ADR64685.1 AGBW02008878 OWR52312.1 KU702627 AMM70654.1 NWSH01004502 PCG65035.1 KY225535 ARO70547.1 KF487731 AII01129.1 KQ459232 KPJ02802.1 KQ461155 KPJ08135.1 KP975110 ALT31629.1 KT381535 ALR72541.1 KU291857 AOE48107.1 APCN01004347 KY283575 AST36235.1 LC017789 BAR64803.1 MG820670 AXF48841.1 PCG65036.1 KY283704 AST36363.1 AXCM01010715 KY325455 AQM73616.1 KQ971338 KYB28047.1 JXUM01071488 KQ562666 KXJ75363.1 CH477363 EAT42606.1 JN836716 AFC91756.2 AAAB01008960 EGK97276.1 KX298830 APC94347.1 EAA11187.5 CVRI01000054 CRL00245.1 KX609444 KB465935 ARN17851.1 RLZ02208.1 KQ971372 EFA10663.2 KP296776 AJO62240.1 KB632161 ERL89256.1 APGK01043551 APGK01043552 APGK01043553 KB741015 ENN75306.1 GENK01000124 JAV45789.1 KY019178 ARB05667.1 KY225540 ARO70552.1 KY817085 AVH87301.1 ATLV01019703 KE525278 KFB44546.1 SOQ43533.1 KX290685 APC94256.1 GGOB01000048 MCH29444.1 KT268301 ANQ46495.1 GEDO01000004 JAP88622.1 KM251708 AKC58589.1 MF975509 AUF73085.1 GCVX01000136 JAI18094.1 KK852952 KDR13328.1 KU702628 AMM70655.1 FX982942 BAU20268.1 KK852854 KDR14986.1 PYGN01000296 PSN49053.1 CH478187 EAT33629.1 PSN49052.1 KX655899 AOG12848.1 PSN49051.1 PYGN01001184 PSN36819.1 JXUM01059078 JXUM01059079 JXUM01059080 JXUM01059081 JXUM01059082 KQ562034 KXJ76850.1 AXCN02000940 NNAY01000335 OXU29125.1 KU702625 AMM70652.1 KX298831 APC94348.1 KY325454 AQM73615.1 NEVH01013550 PNF28810.1 PNF28811.1
Proteomes
UP000283053
UP000007151
UP000218220
UP000053268
UP000053240
UP000076407
+ More
UP000075900 UP000075885 UP000075840 UP000075884 UP000075883 UP000076408 UP000007266 UP000069940 UP000249989 UP000008820 UP000075902 UP000007062 UP000183832 UP000069272 UP000030742 UP000019118 UP000075881 UP000075880 UP000030765 UP000027135 UP000245037 UP000075886 UP000215335 UP000235965
UP000075900 UP000075885 UP000075840 UP000075884 UP000075883 UP000076408 UP000007266 UP000069940 UP000249989 UP000008820 UP000075902 UP000007062 UP000183832 UP000069272 UP000030742 UP000019118 UP000075881 UP000075880 UP000030765 UP000027135 UP000245037 UP000075886 UP000215335 UP000235965
Pfam
Interpro
SUPFAM
SSF81340
SSF81340
Gene 3D
ProteinModelPortal
A0A386H9K5
A0A1W5LAS2
A0A0F7QIG2
A0A2D0WKS5
A0A0S1TQ33
A0A223HCX7
+ More
A0A3S2L3E3 A0A1B3B728 A0A146JYY8 A0A1Q1PP98 A0A2H1VST4 H9A5R2 A0A345BF14 A0A0K8TUD4 A0A223HDB2 E5FIB0 A0A212FEZ5 A0A140G9H5 A0A2A4IYY8 A0A2K8GL75 A0A076E9E7 A0A194QBG6 A0A194QT07 A0A0U3C6J1 A0A0S3J2Z8 A0A1B3B7C4 A0A182XCS1 A0A182RN63 A0A182PN91 A0A182HM47 A0A223HCY5 A0A0F7QIG0 A0A345BF17 A0A2A4IZW1 A0A182NM91 A0A223HDB5 A0A182M3R5 A0A1Q1PP97 A0A182Y0V0 A0A139WJQ1 A0A182GL17 Q178H7 A0A182TLZ3 A0A1S4FBU2 H9A5R6 F5HLW5 A0A1J0KKU9 Q7Q771 A0A1J1IJ30 A0A1W6L166 D6X2Y3 A0A182FFC7 A0A0C5DMP4 U4UF86 N6U3W0 A0A1V1WBQ2 A0A182KER7 A0A2D0WKS7 A0A2K8GL72 A0A182J1B5 A0A2P9JY95 A0A084W2V2 A0A2H1VRQ6 A0A1J0KKK8 A0A310SDV6 A0A1B1FKH3 A0A146JVT1 A0A0E3Y709 A0A2H4ZBE9 A0A0K8TUC6 A0A067R2S3 A0A140G9H6 A0A0U5APN7 A0A067QZN3 A0A2P8YXS4 Q16HB2 A0A2P8YXT7 A0A1B3P5F9 A0A2P8YXT1 A0A2P8XXS9 A0A182GF22 A0A182QH81 A0A232FEK6 A0A140G9H3 A0A1J0KL22 A0A1Q1PP74 A0A182FF56 A0A2J7QJP5 A0A2J7QJP1
A0A3S2L3E3 A0A1B3B728 A0A146JYY8 A0A1Q1PP98 A0A2H1VST4 H9A5R2 A0A345BF14 A0A0K8TUD4 A0A223HDB2 E5FIB0 A0A212FEZ5 A0A140G9H5 A0A2A4IYY8 A0A2K8GL75 A0A076E9E7 A0A194QBG6 A0A194QT07 A0A0U3C6J1 A0A0S3J2Z8 A0A1B3B7C4 A0A182XCS1 A0A182RN63 A0A182PN91 A0A182HM47 A0A223HCY5 A0A0F7QIG0 A0A345BF17 A0A2A4IZW1 A0A182NM91 A0A223HDB5 A0A182M3R5 A0A1Q1PP97 A0A182Y0V0 A0A139WJQ1 A0A182GL17 Q178H7 A0A182TLZ3 A0A1S4FBU2 H9A5R6 F5HLW5 A0A1J0KKU9 Q7Q771 A0A1J1IJ30 A0A1W6L166 D6X2Y3 A0A182FFC7 A0A0C5DMP4 U4UF86 N6U3W0 A0A1V1WBQ2 A0A182KER7 A0A2D0WKS7 A0A2K8GL72 A0A182J1B5 A0A2P9JY95 A0A084W2V2 A0A2H1VRQ6 A0A1J0KKK8 A0A310SDV6 A0A1B1FKH3 A0A146JVT1 A0A0E3Y709 A0A2H4ZBE9 A0A0K8TUC6 A0A067R2S3 A0A140G9H6 A0A0U5APN7 A0A067QZN3 A0A2P8YXS4 Q16HB2 A0A2P8YXT7 A0A1B3P5F9 A0A2P8YXT1 A0A2P8XXS9 A0A182GF22 A0A182QH81 A0A232FEK6 A0A140G9H3 A0A1J0KL22 A0A1Q1PP74 A0A182FF56 A0A2J7QJP5 A0A2J7QJP1
PDB
6NJN
E-value=2.15828e-10,
Score=159
Ontologies
GO
Topology
Subcellular location
Membrane
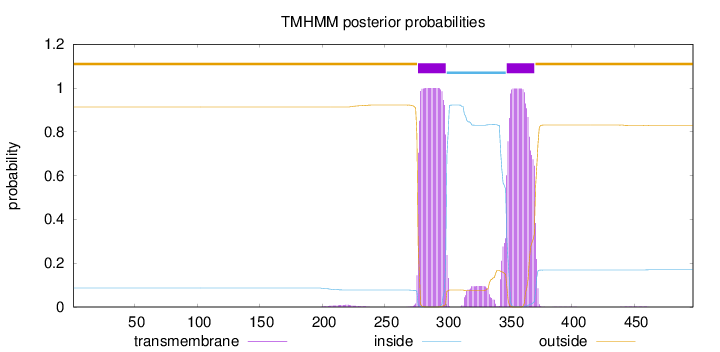
Length:
497
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
47.2024600000001
Exp number, first 60 AAs:
0.00014
Total prob of N-in:
0.08649
outside
1 - 276
TMhelix
277 - 299
inside
300 - 347
TMhelix
348 - 370
outside
371 - 497
Population Genetic Test Statistics
Pi
185.54107
Theta
164.231534
Tajima's D
0.470488
CLR
0.252065
CSRT
0.509424528773561
Interpretation
Uncertain
