Gene
KWMTBOMO10269
Annotation
PREDICTED:_uncharacterized_protein_LOC106129568_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 3.537
Sequence
CDS
ATGCCGATCACCAGGAGCAACAAGGGATCATCGAGGGAACAGCCACTCGAACCTGCGGTTCAAGAATCCAGTCCGGAGGCAACAGAAGCGACGTCGTACGAAACATACGAGAGCTCCGCGACCGGTACGACCCAGAACACAAAACTTGAAGAGGCAACGACGCGGACACGTGGTAAAAATAGACCCGCGTCGAAGAAATCGCTACGACGTAAGGAACTGGCAGCGAGAGCGGAGCTGGCGGAAAAAGAATTGATAGAAGCACAAGCAGCCACCATAGCCGCCAGACTCCGCCTGCAATTGATTCAGGCCGAAAGCGAAGACGACGATGATTCCATTATAGAAGAGCCAAAGGACCGGGAGGACCAGGTCAGGGCCTGGGTCCAGACGTCGGTGGGCAGTATGGACAAGGAGCCCGCAGGCGAGCACATTGAGAACCGTCCCGTCAGCAAGGCCAACATGGAAGAGTGCGGCAACAGGGACCTGGCTAAGGCGATAGTGGAAGCAGTAAGGAGCGCAACGACCAAGGAGCTGCCGCCACAACCCGACTACATGCACCAGTTGCCGACGTTCGAAGGCGACTGCAGGGAGTGGGTGGCCTTCAAGGCTGTCTACGAAGATACCGCTCCACTCTTCTCAAGCGCTCAAAACATGGCGCGGCTGAGAAGAGCGCTGAAGGGACAAGCCAGGGAAGGAGTCCGCAGCCAGCTGTACTCCGAGAGCACACCGGAGGAAGTAATGGAATCACTGCGGCGGTGCTATGGAAGGGCGGACGCTTTAGTCTTAGCGGAGCTAGACCGCATACGAAGGCTTCCAACGATAAGTGACTCCGCCTACGACATTTGCTCACTGGCGAGCAATGTTAACAATACCGTCGCAGCGATCAGGGGACTGCGGAAACCCCAATATTTGATGTCGCCCGAACTGCTGAAGAGCATCGTCGACAAGATGCCGACCATCCTGAGGTTCCGGTGGTACGATCACATTATCGAAAAGACTGAAGGAGACCCGGACCTCAACACGATGTGCGGCTTCCTGAACAAGATGGCGGACCGATGCGGGCCACACGCCGTGGTTGAAGTAGCAACGAAAAGACAACCATCGAGGCGACAGGCGACGCACGCCACAACCGACTGCACCGAGCGTGCAACTTCGGACAGAGCAATGAGGGACAAGCCGGCGTGCCCGCAATGCAAGGATGGACACCCACTATACGACTGTCCGAGGTACAAGGATGCTACGGTGCAGGAACGATGGAGAATGGCAAAGGAGTCGCGCCTGTGTTATAAGTGCCTCGACGGTAAGCACCGGAAGGAAACATGTCAACGACCGCCATGCCGCACGTGCAAGCGCTGGCACCACTACTCGCTACACAACGAGAAGCCGGAGACAGAGCCAGCAGAGAACAGGGAAATCGCGAACAACGTGCGACTTCCCATCAACTGCACACGGATCAGCAGGGCGTACTTGAAGATGGTGCCCGTTGAAGTCTACGGGCCATCGGGATCGACGACGGTGCTGGCCCTGCTGGACGAAGGCTCCACGGTAACTCTCCTGGACTCGAGCGTGGCGAAGCAGATCGGCCTGAAGGGACCCAAGGAGACGCTCCGCCTTGAGACCGTGGGCGGGAAGACCCTAACGAAGAAGGACTCGATGAAGTTTGACATCCGAGTTCGAGGGCTTCACCAGCGGCAGAAGAGGACGATCGTCGGAGCGCGCACAATGGATGACATGAGCCTTGCACCCCAGCGACTAAAGAGGGAAACCATCGAAGGCTACGCGCACCTGAAAAGCCTGGCGGACCAGTTATGTTACGATGAACAATCCCCCCAACTCTTGATCGGGCAGGACAACTGGGGACTCATCGCCGCCAAGACGACGCGCAGGGGCAGGAGGAACCAGCCGGTAGCCTCATTCACCCAATTGGGATGGGTGCTACATGGTCTCGACGGCAACAACGCAAAAGAGGTCAACTTCCTCACCTGCGCCCATGTATCGATGCGCGAAGAGGCCTTGGACAGACTGGTGAAGGAGCACTTCACGATAGAGTCCCACCCAATCGTCCGAGCACGGATATGGAGGCGAAGGCGTTTGACATCTTGTAGAAGGAGGACGACAGTGGTTGGAGACGCGATGACGGCCGCTTCGAAGCTCGGACCTACAATCCCTGTTCGAAGCGTTGTTGCGGTTCCGAAAAAGGCTCGTCATGGTTGTCGCGGACATCAAGGAGAGGTTCCTGCAGATAAGGATCAGGGACGAGGACCGCGACAGCCTGAGATTCCTCTATAG
Protein
MPITRSNKGSSREQPLEPAVQESSPEATEATSYETYESSATGTTQNTKLEEATTRTRGKNRPASKKSLRRKELAARAELAEKELIEAQAATIAARLRLQLIQAESEDDDDSIIEEPKDREDQVRAWVQTSVGSMDKEPAGEHIENRPVSKANMEECGNRDLAKAIVEAVRSATTKELPPQPDYMHQLPTFEGDCREWVAFKAVYEDTAPLFSSAQNMARLRRALKGQAREGVRSQLYSESTPEEVMESLRRCYGRADALVLAELDRIRRLPTISDSAYDICSLASNVNNTVAAIRGLRKPQYLMSPELLKSIVDKMPTILRFRWYDHIIEKTEGDPDLNTMCGFLNKMADRCGPHAVVEVATKRQPSRRQATHATTDCTERATSDRAMRDKPACPQCKDGHPLYDCPRYKDATVQERWRMAKESRLCYKCLDGKHRKETCQRPPCRTCKRWHHYSLHNEKPETEPAENREIANNVRLPINCTRISRAYLKMVPVEVYGPSGSTTVLALLDEGSTVTLLDSSVAKQIGLKGPKETLRLETVGGKTLTKKDSMKFDIRVRGLHQRQKRTIVGARTMDDMSLAPQRLKRETIEGYAHLKSLADQLCYDEQSPQLLIGQDNWGLIAAKTTRRGRRNQPVASFTQLGWVLHGLDGNNAKEVNFLTCAHVSMREEALDRLVKEHFTIESHPIVRARIWRRRRLTSCRRRTTVVGDAMTAASKLGPTIPVRSVVAVPKKARHGCRGHQGEVPADKDQGRGPRQPEIPL
Summary
Uniprot
Q2MGA5
A0A2H1W036
V9GZQ1
A0A3S2N5J7
A0A2A4IX21
H9JNE5
+ More
A0A0L7KW57 A0A0L7L0N3 A0A0A9ZAM6 A0A0A9ZJ56 A0A0A9ZFI6 A0A0L7KYG1 A0A0A9ZGM2 A0A0A9ZDG1 A0A182H5B8 A0A182GC70 A0A226DH22 A0A182HDB0 A0A226E2P7 A0A226EEI4 A0A226EDU9 A0A182RWX5 Q76C94 Q76C95 O02006 A0A182HEF9 A0A2M4CKZ8 A0A1L8DIN5 A0A1L8DJ10 A0A226D278 A0A226EKF8 A0A1W7R6D5 A0A1W7R699
A0A0L7KW57 A0A0L7L0N3 A0A0A9ZAM6 A0A0A9ZJ56 A0A0A9ZFI6 A0A0L7KYG1 A0A0A9ZGM2 A0A0A9ZDG1 A0A182H5B8 A0A182GC70 A0A226DH22 A0A182HDB0 A0A226E2P7 A0A226EEI4 A0A226EDU9 A0A182RWX5 Q76C94 Q76C95 O02006 A0A182HEF9 A0A2M4CKZ8 A0A1L8DIN5 A0A1L8DJ10 A0A226D278 A0A226EKF8 A0A1W7R6D5 A0A1W7R699
EMBL
AF530470
AAQ09229.1
ODYU01005530
SOQ46470.1
L09635
AAA75005.1
+ More
RSAL01000375 RVE42106.1 NWSH01005440 PCG64209.1 BABH01019236 JTDY01005167 KOB67301.1 JTDY01003744 KOB69058.1 GBHO01002130 JAG41474.1 GBHO01000309 JAG43295.1 GBHO01000310 JAG43294.1 JTDY01004423 KOB68175.1 GBHO01000308 JAG43296.1 GBHO01000307 JAG43297.1 JXUM01111221 KQ565482 KXJ70922.1 JXUM01053894 KQ561796 KXJ77517.1 LNIX01000019 OXA44855.1 JXUM01034199 KQ561015 KXJ80028.1 LNIX01000007 OXA52015.1 LNIX01000004 OXA55241.1 OXA55408.1 AB110069 BAD01590.1 BAD01589.1 D83207 BAA57030.1 JXUM01005879 KQ560203 KXJ83865.1 GGFL01001727 MBW65905.1 GFDF01007772 JAV06312.1 GFDF01007760 JAV06324.1 LNIX01000039 OXA39263.1 LNIX01000003 OXA57597.1 GEHC01000921 JAV46724.1 GEHC01000956 JAV46689.1
RSAL01000375 RVE42106.1 NWSH01005440 PCG64209.1 BABH01019236 JTDY01005167 KOB67301.1 JTDY01003744 KOB69058.1 GBHO01002130 JAG41474.1 GBHO01000309 JAG43295.1 GBHO01000310 JAG43294.1 JTDY01004423 KOB68175.1 GBHO01000308 JAG43296.1 GBHO01000307 JAG43297.1 JXUM01111221 KQ565482 KXJ70922.1 JXUM01053894 KQ561796 KXJ77517.1 LNIX01000019 OXA44855.1 JXUM01034199 KQ561015 KXJ80028.1 LNIX01000007 OXA52015.1 LNIX01000004 OXA55241.1 OXA55408.1 AB110069 BAD01590.1 BAD01589.1 D83207 BAA57030.1 JXUM01005879 KQ560203 KXJ83865.1 GGFL01001727 MBW65905.1 GFDF01007772 JAV06312.1 GFDF01007760 JAV06324.1 LNIX01000039 OXA39263.1 LNIX01000003 OXA57597.1 GEHC01000921 JAV46724.1 GEHC01000956 JAV46689.1
Proteomes
Pfam
Interpro
IPR040676
DUF5641
+ More
IPR036397 RNaseH_sf
IPR005312 DUF1759
IPR001584 Integrase_cat-core
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR001841 Znf_RING
IPR019786 Zinc_finger_PHD-type_CS
IPR000477 RT_dom
IPR019787 Znf_PHD-finger
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR011011 Znf_FYVE_PHD
IPR006094 Oxid_FAD_bind_N
IPR016166 FAD-bd_PCMH
IPR016164 FAD-linked_Oxase-like_C
IPR041588 Integrase_H2C2
IPR016170 Cytok_DH_C_sf
IPR015213 Cholesterol_OX_subst-bd
IPR036318 FAD-bd_PCMH-like_sf
IPR016169 FAD-bd_PCMH_sub2
IPR016167 FAD-bd_PCMH_sub1
IPR008737 Peptidase_asp_put
IPR036397 RNaseH_sf
IPR005312 DUF1759
IPR001584 Integrase_cat-core
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR001841 Znf_RING
IPR019786 Zinc_finger_PHD-type_CS
IPR000477 RT_dom
IPR019787 Znf_PHD-finger
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR011011 Znf_FYVE_PHD
IPR006094 Oxid_FAD_bind_N
IPR016166 FAD-bd_PCMH
IPR016164 FAD-linked_Oxase-like_C
IPR041588 Integrase_H2C2
IPR016170 Cytok_DH_C_sf
IPR015213 Cholesterol_OX_subst-bd
IPR036318 FAD-bd_PCMH-like_sf
IPR016169 FAD-bd_PCMH_sub2
IPR016167 FAD-bd_PCMH_sub1
IPR008737 Peptidase_asp_put
ProteinModelPortal
Q2MGA5
A0A2H1W036
V9GZQ1
A0A3S2N5J7
A0A2A4IX21
H9JNE5
+ More
A0A0L7KW57 A0A0L7L0N3 A0A0A9ZAM6 A0A0A9ZJ56 A0A0A9ZFI6 A0A0L7KYG1 A0A0A9ZGM2 A0A0A9ZDG1 A0A182H5B8 A0A182GC70 A0A226DH22 A0A182HDB0 A0A226E2P7 A0A226EEI4 A0A226EDU9 A0A182RWX5 Q76C94 Q76C95 O02006 A0A182HEF9 A0A2M4CKZ8 A0A1L8DIN5 A0A1L8DJ10 A0A226D278 A0A226EKF8 A0A1W7R6D5 A0A1W7R699
A0A0L7KW57 A0A0L7L0N3 A0A0A9ZAM6 A0A0A9ZJ56 A0A0A9ZFI6 A0A0L7KYG1 A0A0A9ZGM2 A0A0A9ZDG1 A0A182H5B8 A0A182GC70 A0A226DH22 A0A182HDB0 A0A226E2P7 A0A226EEI4 A0A226EDU9 A0A182RWX5 Q76C94 Q76C95 O02006 A0A182HEF9 A0A2M4CKZ8 A0A1L8DIN5 A0A1L8DJ10 A0A226D278 A0A226EKF8 A0A1W7R6D5 A0A1W7R699
Ontologies
GO
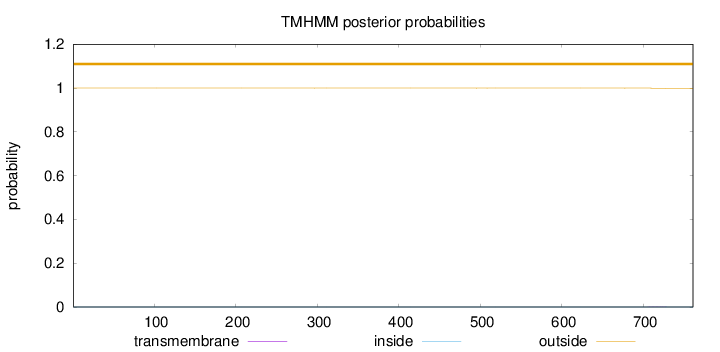
Topology
Length:
761
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00602
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00016
outside
1 - 761
Population Genetic Test Statistics
Pi
177.486752
Theta
138.69884
Tajima's D
0.631989
CLR
253.2628
CSRT
0.551372431378431
Interpretation
Uncertain
