Gene
KWMTBOMO10266
Pre Gene Modal
BGIBMGA007274
Annotation
PREDICTED:_protein_I'm_not_dead_yet-like_[Plutella_xylostella]
Full name
Protein I'm not dead yet
+ More
Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit
Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit
Alternative Name
INDY transporter protein
drIndy
drIndy
Location in the cell
PlasmaMembrane Reliability : 2.743
Sequence
CDS
ATGTTACTTATGGAGTTAGCTATGTATTTTTATTTGATTATCGTGTACTTCGGATTTTTAAGACCCAATAGCCAAGCTGCTAAAGATTCTGTGATCACCCCAGAAAGTATCGAAGCAGCAAAAAAGACCGTTGATAAAGATTGGGTAAATTTGGGAAAAGTAACATTTTGGGAATATATGGTAATCATATTATTTGGTGGAGCAATGGTATTATTCTTTGCAAGATCCCCTCAAATTTTTCAAGGTTGGGGCGATAAAACCATCAGTTATTTCGGGATTAAAAATAACAAATTGTAA
Protein
MLLMELAMYFYLIIVYFGFLRPNSQAAKDSVITPESIEAAKKTVDKDWVNLGKVTFWEYMVIILFGGAMVLFFARSPQIFQGWGDKTISYFGIKNNKL
Summary
Description
Cation-independent electroneutral transporter (not associated with membrane depolarization) of a variety of tricarboxylic and dicarboxylic acid-cycle intermediates. There is also small, but detectable, transport of monocarboxylics. Transport is through the epithelium of the gut and across the plasma membranes of organs involved in intermediary metabolism and storage. Affinity for substrates is citrate > succinate > pyruvate. Fumarate, a-ketoglutarate, and glutarate are also transported, but not lactate. Transport mechanism that is not coupled to Na(+), K(+), or Cl(-). Function is shown in Xenopus oocytes and human retinal pigment epithelial (HRPE) cell lines.
Subunit of the oligosaccharyl transferase (OST) complex that catalyzes the initial transfer of a defined glycan (Glc(3)Man(9)GlcNAc(2) in eukaryotes) from the lipid carrier dolichol-pyrophosphate to an asparagine residue within an Asn-X-Ser/Thr consensus motif in nascent polypeptide chains, the first step in protein N-glycosylation. N-glycosylation occurs cotranslationally and the complex associates with the Sec61 complex at the channel-forming translocon complex that mediates protein translocation across the endoplasmic reticulum (ER).
Subunit of the oligosaccharyl transferase (OST) complex that catalyzes the initial transfer of a defined glycan (Glc(3)Man(9)GlcNAc(2) in eukaryotes) from the lipid carrier dolichol-pyrophosphate to an asparagine residue within an Asn-X-Ser/Thr consensus motif in nascent polypeptide chains, the first step in protein N-glycosylation. N-glycosylation occurs cotranslationally and the complex associates with the Sec61 complex at the channel-forming translocon complex that mediates protein translocation across the endoplasmic reticulum (ER).
Subunit
Component of the oligosaccharyltransferase (OST) complex.
Miscellaneous
The life-extending effect of mutations is likely caused by an alteration in energy balance caused by a decrease in transport function.
Similarity
Belongs to the SLC13A/DASS transporter (TC 2.A.47) family. NADC subfamily.
Belongs to the DDOST 48 kDa subunit family.
Belongs to the DDOST 48 kDa subunit family.
Keywords
Alternative splicing
Cell membrane
Complete proteome
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Transport
Feature
chain Protein I'm not dead yet
splice variant In isoform B.
splice variant In isoform B.
Uniprot
A0A3S2NWJ4
A0A212FAN4
A0A194QRQ2
A0A2A4J2F5
A0A3S2NFT2
A0A2A4JSL5
+ More
A0A194QH25 A0A2H1VD01 T1GDJ2 A0A182GSH0 W5J711 A0A2M3ZG83 A0A1L8DEL7 A0A1L8DES5 A0A1L8DE76 A0A1L8DE04 A0A182GSH2 A0A182FER4 A0A1B0GFN7 A0A1B0A737 A0A336KEI2 A0A1A9V5X8 A0A336KWG5 A0A336KDB7 A0A182SQS9 Q16UM1 A0A1L8ECM3 A0A1L8E9V4 A0A0P6JNM3 A0A1S4FNK3 Q16UM2 A0A182NZ46 A0A182WWT0 Q7QIT2 A0A182VBY4 A0A182L392 A0A182UAV5 A0A182HWF9 B4J1T3 A0A182GSH1 A0A1A9XD94 A0A1B0C4D6 A0A182WL11 A0A182JX44 A0A182Y266 A0A1A9W3H3 A0A0P8XVW7 B3M868 A0A2M4BIC4 A0A2M4BIG4 A0A182Q6Q6 A0A0L0C679 A0A182JDR1 A0A0C9RKM7 A0A1I8QD75 E9IWV9 A0A182PTK9 A0A1W4WEQ3 B3NHN6 A0A0Q5UJB0 Q9VVT2-2 A0A0J9RXY5 E1JI19 B4IFW0 D0IQJ2 Q9VVT2 A0A0J9RXQ8 A0A0C9QGV0 A0A0R1DXX2 B4QPW6 A0A195CS08 B4PIY5 A0A151WJU9 A0A195E354 B4N4U3 A0A0R3P3N0 A0A182RMY3 B5DPF3 U5EXM5 A0A0Q9XRT0 B4KXS1 A0A154PRT9 A0A084WJU3 Q16UM0 T1PEL6 A0A0M4ETS4 A0A1S4FNY7 A0A1Q3FP35 A0A182J1X2 A0A1Q3FNV3 K7IW72 K7J4H9 A0A1I8M3N9 A0A0Q9WVC4 B4LFX1 A0A0Q9WUJ9 A0A0M4EI95 A0A1J1IEA9
A0A194QH25 A0A2H1VD01 T1GDJ2 A0A182GSH0 W5J711 A0A2M3ZG83 A0A1L8DEL7 A0A1L8DES5 A0A1L8DE76 A0A1L8DE04 A0A182GSH2 A0A182FER4 A0A1B0GFN7 A0A1B0A737 A0A336KEI2 A0A1A9V5X8 A0A336KWG5 A0A336KDB7 A0A182SQS9 Q16UM1 A0A1L8ECM3 A0A1L8E9V4 A0A0P6JNM3 A0A1S4FNK3 Q16UM2 A0A182NZ46 A0A182WWT0 Q7QIT2 A0A182VBY4 A0A182L392 A0A182UAV5 A0A182HWF9 B4J1T3 A0A182GSH1 A0A1A9XD94 A0A1B0C4D6 A0A182WL11 A0A182JX44 A0A182Y266 A0A1A9W3H3 A0A0P8XVW7 B3M868 A0A2M4BIC4 A0A2M4BIG4 A0A182Q6Q6 A0A0L0C679 A0A182JDR1 A0A0C9RKM7 A0A1I8QD75 E9IWV9 A0A182PTK9 A0A1W4WEQ3 B3NHN6 A0A0Q5UJB0 Q9VVT2-2 A0A0J9RXY5 E1JI19 B4IFW0 D0IQJ2 Q9VVT2 A0A0J9RXQ8 A0A0C9QGV0 A0A0R1DXX2 B4QPW6 A0A195CS08 B4PIY5 A0A151WJU9 A0A195E354 B4N4U3 A0A0R3P3N0 A0A182RMY3 B5DPF3 U5EXM5 A0A0Q9XRT0 B4KXS1 A0A154PRT9 A0A084WJU3 Q16UM0 T1PEL6 A0A0M4ETS4 A0A1S4FNY7 A0A1Q3FP35 A0A182J1X2 A0A1Q3FNV3 K7IW72 K7J4H9 A0A1I8M3N9 A0A0Q9WVC4 B4LFX1 A0A0Q9WUJ9 A0A0M4EI95 A0A1J1IEA9
Pubmed
22118469
26354079
26483478
20920257
23761445
17510324
+ More
12364791 14747013 17210077 20966253 17994087 25244985 26108605 21282665 12186628 10731132 12537572 12537569 10581279 11118146 12391301 22936249 12537568 12537573 12537574 16110336 17569856 17569867 17550304 15632085 24438588 20075255 25315136
12364791 14747013 17210077 20966253 17994087 25244985 26108605 21282665 12186628 10731132 12537572 12537569 10581279 11118146 12391301 22936249 12537568 12537573 12537574 16110336 17569856 17569867 17550304 15632085 24438588 20075255 25315136
EMBL
RSAL01000130
RVE46445.1
AGBW02009440
OWR50804.1
KQ461155
KPJ08193.1
+ More
NWSH01003418 PCG66337.1 RVE46444.1 NWSH01000682 PCG74819.1 KQ459232 KPJ02746.1 ODYU01001889 SOQ38729.1 CAQQ02394832 CAQQ02394833 JXUM01002644 KQ560143 KXJ84194.1 ADMH02002049 ETN59761.1 GGFM01006709 MBW27460.1 GFDF01009206 JAV04878.1 GFDF01009207 JAV04877.1 GFDF01009399 JAV04685.1 GFDF01009398 JAV04686.1 JXUM01002648 KXJ84196.1 CCAG010022811 UFQS01000322 UFQT01000322 SSX02867.1 SSX23234.1 UFQS01001198 UFQT01001198 SSX09649.1 SSX29445.1 SSX02866.1 SSX23233.1 CH477618 EAT38222.1 GFDG01002319 JAV16480.1 GFDG01003239 JAV15560.1 GDIQ01002285 JAN92452.1 EAT38221.1 AAAB01008807 EAA03999.4 APCN01001489 CH916366 EDV98013.1 JXUM01002647 KXJ84195.1 JXJN01025422 CH902618 KPU78880.1 EDV41007.1 GGFJ01003664 MBW52805.1 GGFJ01003663 MBW52804.1 AXCN02000497 JRES01000836 KNC27883.1 GBYB01013822 JAG83589.1 GL766616 EFZ14935.1 CH954178 EDV51831.1 KQS43994.1 AF509505 AE014296 AY102686 AF217399 AAF73384.1 AAM27515.1 CM002912 KMZ00448.1 ACZ94736.1 CH480834 EDW46547.1 BT100230 ACY56898.1 KMZ00449.1 GBYB01013823 JAG83590.1 CM000159 KRK01936.1 CM000363 EDX10994.1 KQ977329 KYN03523.1 EDW94576.1 KQ983028 KYQ48153.1 KQ979701 KYN19511.1 CH964101 EDW79382.1 CH379069 KRT07809.1 EDY73532.1 GANO01002475 JAB57396.1 CH933809 KRG06824.1 EDW19778.1 KQ435090 KZC14611.1 ATLV01024071 ATLV01024072 KE525348 KFB50487.1 EAT38223.1 KA647227 AFP61856.1 CP012526 ALC46330.1 GFDL01005799 JAV29246.1 GFDL01005766 JAV29279.1 CH940647 KRF84878.1 EDW70370.1 KRF84879.1 CP012525 ALC44427.1 CVRI01000047 CRK98547.1
NWSH01003418 PCG66337.1 RVE46444.1 NWSH01000682 PCG74819.1 KQ459232 KPJ02746.1 ODYU01001889 SOQ38729.1 CAQQ02394832 CAQQ02394833 JXUM01002644 KQ560143 KXJ84194.1 ADMH02002049 ETN59761.1 GGFM01006709 MBW27460.1 GFDF01009206 JAV04878.1 GFDF01009207 JAV04877.1 GFDF01009399 JAV04685.1 GFDF01009398 JAV04686.1 JXUM01002648 KXJ84196.1 CCAG010022811 UFQS01000322 UFQT01000322 SSX02867.1 SSX23234.1 UFQS01001198 UFQT01001198 SSX09649.1 SSX29445.1 SSX02866.1 SSX23233.1 CH477618 EAT38222.1 GFDG01002319 JAV16480.1 GFDG01003239 JAV15560.1 GDIQ01002285 JAN92452.1 EAT38221.1 AAAB01008807 EAA03999.4 APCN01001489 CH916366 EDV98013.1 JXUM01002647 KXJ84195.1 JXJN01025422 CH902618 KPU78880.1 EDV41007.1 GGFJ01003664 MBW52805.1 GGFJ01003663 MBW52804.1 AXCN02000497 JRES01000836 KNC27883.1 GBYB01013822 JAG83589.1 GL766616 EFZ14935.1 CH954178 EDV51831.1 KQS43994.1 AF509505 AE014296 AY102686 AF217399 AAF73384.1 AAM27515.1 CM002912 KMZ00448.1 ACZ94736.1 CH480834 EDW46547.1 BT100230 ACY56898.1 KMZ00449.1 GBYB01013823 JAG83590.1 CM000159 KRK01936.1 CM000363 EDX10994.1 KQ977329 KYN03523.1 EDW94576.1 KQ983028 KYQ48153.1 KQ979701 KYN19511.1 CH964101 EDW79382.1 CH379069 KRT07809.1 EDY73532.1 GANO01002475 JAB57396.1 CH933809 KRG06824.1 EDW19778.1 KQ435090 KZC14611.1 ATLV01024071 ATLV01024072 KE525348 KFB50487.1 EAT38223.1 KA647227 AFP61856.1 CP012526 ALC46330.1 GFDL01005799 JAV29246.1 GFDL01005766 JAV29279.1 CH940647 KRF84878.1 EDW70370.1 KRF84879.1 CP012525 ALC44427.1 CVRI01000047 CRK98547.1
Proteomes
UP000283053
UP000007151
UP000053240
UP000218220
UP000053268
UP000015102
+ More
UP000069940 UP000249989 UP000000673 UP000069272 UP000092444 UP000092445 UP000078200 UP000075901 UP000008820 UP000075884 UP000076407 UP000007062 UP000075903 UP000075882 UP000075902 UP000075840 UP000001070 UP000092443 UP000092460 UP000075920 UP000075881 UP000076408 UP000091820 UP000007801 UP000075886 UP000037069 UP000075880 UP000095300 UP000075885 UP000192223 UP000008711 UP000000803 UP000001292 UP000002282 UP000000304 UP000078542 UP000075809 UP000078492 UP000007798 UP000001819 UP000075900 UP000009192 UP000076502 UP000030765 UP000092553 UP000002358 UP000095301 UP000008792 UP000183832
UP000069940 UP000249989 UP000000673 UP000069272 UP000092444 UP000092445 UP000078200 UP000075901 UP000008820 UP000075884 UP000076407 UP000007062 UP000075903 UP000075882 UP000075902 UP000075840 UP000001070 UP000092443 UP000092460 UP000075920 UP000075881 UP000076408 UP000091820 UP000007801 UP000075886 UP000037069 UP000075880 UP000095300 UP000075885 UP000192223 UP000008711 UP000000803 UP000001292 UP000002282 UP000000304 UP000078542 UP000075809 UP000078492 UP000007798 UP000001819 UP000075900 UP000009192 UP000076502 UP000030765 UP000092553 UP000002358 UP000095301 UP000008792 UP000183832
Pfam
Interpro
SUPFAM
SSF57610
SSF57610
Gene 3D
CDD
ProteinModelPortal
A0A3S2NWJ4
A0A212FAN4
A0A194QRQ2
A0A2A4J2F5
A0A3S2NFT2
A0A2A4JSL5
+ More
A0A194QH25 A0A2H1VD01 T1GDJ2 A0A182GSH0 W5J711 A0A2M3ZG83 A0A1L8DEL7 A0A1L8DES5 A0A1L8DE76 A0A1L8DE04 A0A182GSH2 A0A182FER4 A0A1B0GFN7 A0A1B0A737 A0A336KEI2 A0A1A9V5X8 A0A336KWG5 A0A336KDB7 A0A182SQS9 Q16UM1 A0A1L8ECM3 A0A1L8E9V4 A0A0P6JNM3 A0A1S4FNK3 Q16UM2 A0A182NZ46 A0A182WWT0 Q7QIT2 A0A182VBY4 A0A182L392 A0A182UAV5 A0A182HWF9 B4J1T3 A0A182GSH1 A0A1A9XD94 A0A1B0C4D6 A0A182WL11 A0A182JX44 A0A182Y266 A0A1A9W3H3 A0A0P8XVW7 B3M868 A0A2M4BIC4 A0A2M4BIG4 A0A182Q6Q6 A0A0L0C679 A0A182JDR1 A0A0C9RKM7 A0A1I8QD75 E9IWV9 A0A182PTK9 A0A1W4WEQ3 B3NHN6 A0A0Q5UJB0 Q9VVT2-2 A0A0J9RXY5 E1JI19 B4IFW0 D0IQJ2 Q9VVT2 A0A0J9RXQ8 A0A0C9QGV0 A0A0R1DXX2 B4QPW6 A0A195CS08 B4PIY5 A0A151WJU9 A0A195E354 B4N4U3 A0A0R3P3N0 A0A182RMY3 B5DPF3 U5EXM5 A0A0Q9XRT0 B4KXS1 A0A154PRT9 A0A084WJU3 Q16UM0 T1PEL6 A0A0M4ETS4 A0A1S4FNY7 A0A1Q3FP35 A0A182J1X2 A0A1Q3FNV3 K7IW72 K7J4H9 A0A1I8M3N9 A0A0Q9WVC4 B4LFX1 A0A0Q9WUJ9 A0A0M4EI95 A0A1J1IEA9
A0A194QH25 A0A2H1VD01 T1GDJ2 A0A182GSH0 W5J711 A0A2M3ZG83 A0A1L8DEL7 A0A1L8DES5 A0A1L8DE76 A0A1L8DE04 A0A182GSH2 A0A182FER4 A0A1B0GFN7 A0A1B0A737 A0A336KEI2 A0A1A9V5X8 A0A336KWG5 A0A336KDB7 A0A182SQS9 Q16UM1 A0A1L8ECM3 A0A1L8E9V4 A0A0P6JNM3 A0A1S4FNK3 Q16UM2 A0A182NZ46 A0A182WWT0 Q7QIT2 A0A182VBY4 A0A182L392 A0A182UAV5 A0A182HWF9 B4J1T3 A0A182GSH1 A0A1A9XD94 A0A1B0C4D6 A0A182WL11 A0A182JX44 A0A182Y266 A0A1A9W3H3 A0A0P8XVW7 B3M868 A0A2M4BIC4 A0A2M4BIG4 A0A182Q6Q6 A0A0L0C679 A0A182JDR1 A0A0C9RKM7 A0A1I8QD75 E9IWV9 A0A182PTK9 A0A1W4WEQ3 B3NHN6 A0A0Q5UJB0 Q9VVT2-2 A0A0J9RXY5 E1JI19 B4IFW0 D0IQJ2 Q9VVT2 A0A0J9RXQ8 A0A0C9QGV0 A0A0R1DXX2 B4QPW6 A0A195CS08 B4PIY5 A0A151WJU9 A0A195E354 B4N4U3 A0A0R3P3N0 A0A182RMY3 B5DPF3 U5EXM5 A0A0Q9XRT0 B4KXS1 A0A154PRT9 A0A084WJU3 Q16UM0 T1PEL6 A0A0M4ETS4 A0A1S4FNY7 A0A1Q3FP35 A0A182J1X2 A0A1Q3FNV3 K7IW72 K7J4H9 A0A1I8M3N9 A0A0Q9WVC4 B4LFX1 A0A0Q9WUJ9 A0A0M4EI95 A0A1J1IEA9
Ontologies
GO
PANTHER
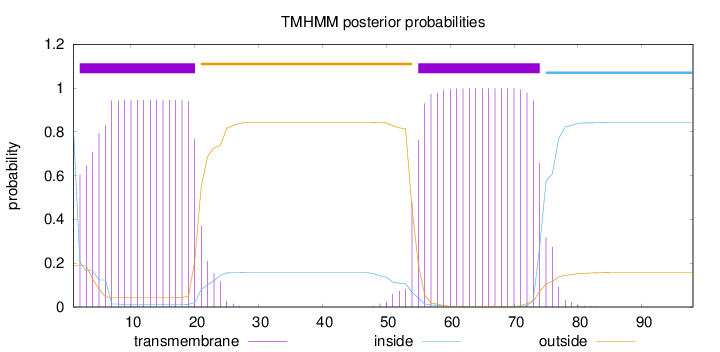
Topology
Subcellular location
Basolateral cell membrane
Endoplasmic reticulum membrane
Endoplasmic reticulum membrane
Length:
98
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
38.20867
Exp number, first 60 AAs:
23.88767
Total prob of N-in:
0.81067
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 20
outside
21 - 54
TMhelix
55 - 74
inside
75 - 98
Population Genetic Test Statistics
Pi
107.675009
Theta
117.662467
Tajima's D
-0.301801
CLR
0.286059
CSRT
0.292235388230588
Interpretation
Uncertain
