Gene
KWMTBOMO10260
Pre Gene Modal
BGIBMGA007276
Annotation
putative_coiled-coil_domain-containing_protein_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 2.586
Sequence
CDS
ATGGTTTTAAAACGTAAACTGGATCATGTTTTAAAACAACTAGACGGAGCTGCAGATTTGGAGGAAAAGTACCAACAAAAGCTATTTTCGTTTGAAGAACTTAAAGAAAAGTCGGAAAGAGAAATAGCGGATATCAAAGCAAAAGTCCAAACTGTACAGCTGCAATACGCGGATCGTTTAGAACAAGCCGAAAAAAAATTTGACGAATTGGAGAACTACGAGAAGAATACAGGAACAGGCCTAATTTATTCAAAAAAGGGCAAACCCATTGCTGACAAGACAGTTCAGAGGTTTTTAACGCTTCAAAGACGCAAATGCGAGCAGTCCTCAGCACTATGCCTCCGTTATATTCGTGCGAGAAACGCTGTGGCTGAACTTGAAGCTATAGTAAGAAAACTCGAGATGCTCGGGCCCGGCTTATACGTGGCTCAATACGAACAATTGGACATCGACCATCAAAATTATATGACAAAAATCGAAGGTGATATTTGTTAA
Protein
MVLKRKLDHVLKQLDGAADLEEKYQQKLFSFEELKEKSEREIADIKAKVQTVQLQYADRLEQAEKKFDELENYEKNTGTGLIYSKKGKPIADKTVQRFLTLQRRKCEQSSALCLRYIRARNAVAELEAIVRKLEMLGPGLYVAQYEQLDIDHQNYMTKIEGDIC
Summary
Uniprot
H9JCN0
A0A2H1WY89
A0A212FAQ0
A0A2A4JRZ8
A0A194QB24
A0A194QT66
+ More
A0A0L7L3F1 E0VNS4 A0A1Y1KVV6 A0A1Y1KSC0 A0A2H1C9E3 A0A1S8WRZ2 H2KVV4 A0A075A2R1 A0A3R7FDP8 A0A183JBY3 F4PFY6 A0A383WPS1 A0A183NUU2 A0A139AUA1 F4PAA1 A7S8B1 A0A177WWW9 A0A195CUY8 A0A183LES3 T2MC62 A0A369S2I1 A0A094ZES1 A0A139WKX7 A0A177WXM0 A0A3M6UM28 A0A151XFY8 G4VBV7 A0A3Q0K433 A0A3Q0KCL4 A0A1W2W8S4 A0A3Q0K1H9 W4XZK1 A0A139WDP5 A0A183RSH0 A0A2B4RTF4 A0A1B6DRU8 A0A1Y1VFA7 A0A1B6CFK9 A0A151IVW0 A0A2V0P647 A0A195FNU1 A0A1B6DLU2 C7TZG2 W5NNC8 A0A060Y5A5 A0A1Y2HPI9 A0A183AAR5 A0A397DBX2 A0A3R7WKB3 A0A397D814 A0A1S8W9A1 G3VZN1 A0A1Y3NND5 A0A3P6P9G5 A0A1S8W9A2 K1RPB0 A0A1S8W9A8 A0A1Y1XNV4 A0A2R5G1A7 A0A2G8JHZ2 F6SBS0 A0A3P8Y293 A0A1Y2CH53 A0A075B3E6 A0A1L8HKU2 A0A3F2RRV2 D6WDQ7 H2ZR54 A0A195BJ87 E9J3L0 D2VA28 A0A061QYD0 A0A061RN40 V9ENT3 A0A0W8BVL1 W2YVK0 W2GFK7 A0A0W8DHT1 W2WK06 A0A329S8J5 L1J9H4 A0A078B1C9 W2R600 W2IJX0 J9I7B9 A0A1Y2C8H9 K3WVZ3
A0A0L7L3F1 E0VNS4 A0A1Y1KVV6 A0A1Y1KSC0 A0A2H1C9E3 A0A1S8WRZ2 H2KVV4 A0A075A2R1 A0A3R7FDP8 A0A183JBY3 F4PFY6 A0A383WPS1 A0A183NUU2 A0A139AUA1 F4PAA1 A7S8B1 A0A177WWW9 A0A195CUY8 A0A183LES3 T2MC62 A0A369S2I1 A0A094ZES1 A0A139WKX7 A0A177WXM0 A0A3M6UM28 A0A151XFY8 G4VBV7 A0A3Q0K433 A0A3Q0KCL4 A0A1W2W8S4 A0A3Q0K1H9 W4XZK1 A0A139WDP5 A0A183RSH0 A0A2B4RTF4 A0A1B6DRU8 A0A1Y1VFA7 A0A1B6CFK9 A0A151IVW0 A0A2V0P647 A0A195FNU1 A0A1B6DLU2 C7TZG2 W5NNC8 A0A060Y5A5 A0A1Y2HPI9 A0A183AAR5 A0A397DBX2 A0A3R7WKB3 A0A397D814 A0A1S8W9A1 G3VZN1 A0A1Y3NND5 A0A3P6P9G5 A0A1S8W9A2 K1RPB0 A0A1S8W9A8 A0A1Y1XNV4 A0A2R5G1A7 A0A2G8JHZ2 F6SBS0 A0A3P8Y293 A0A1Y2CH53 A0A075B3E6 A0A1L8HKU2 A0A3F2RRV2 D6WDQ7 H2ZR54 A0A195BJ87 E9J3L0 D2VA28 A0A061QYD0 A0A061RN40 V9ENT3 A0A0W8BVL1 W2YVK0 W2GFK7 A0A0W8DHT1 W2WK06 A0A329S8J5 L1J9H4 A0A078B1C9 W2R600 W2IJX0 J9I7B9 A0A1Y2C8H9 K3WVZ3
Pubmed
EMBL
BABH01034607
BABH01034608
ODYU01011546
SOQ57354.1
AGBW02009440
OWR50801.1
+ More
NWSH01000682 PCG74817.1 KQ459232 KPJ02743.1 KQ461155 KPJ08195.1 JTDY01003165 KOB70028.1 DS235346 EEB15030.1 GEZM01075064 JAV64310.1 GEZM01075065 JAV64309.1 KZ430352 PIS84031.1 KV895655 OON17252.1 DF145157 GAA38980.2 KL596670 KER29860.1 NIRI01001908 RJW64567.1 UZAK01000104 VDO60005.1 GL882981 EGF75858.1 FNXT01001367 SZX79458.1 UZAL01027264 VDP30882.1 KQ965736 KXS20284.1 GL882890 EGF78030.1 DS469597 EDO40032.1 DS022311 OAJ44134.1 KQ977279 KYN04337.1 UZAI01000574 VDO54487.1 HAAD01003606 CDG69838.1 NOWV01000096 RDD40624.1 KL250496 KGB32247.1 KQ971322 KYB28600.1 OAJ44130.1 RCHS01001219 RMX54659.1 KQ982182 KYQ59225.1 HE601625 CCD77381.1 AAGJ04109380 AAGJ04109381 KQ971358 KYB25987.1 LSMT01000358 PFX19502.1 GEDC01008904 GEDC01008453 JAS28394.1 JAS28845.1 MCFH01000010 ORX54783.1 GEDC01025173 JAS12125.1 KQ980880 KYN11991.1 BDRX01000033 GBF92555.1 KQ981382 KYN42081.1 GEDC01010680 JAS26618.1 FN327264 CAX82988.1 AHAT01001233 FR907431 CDQ86682.1 MCFL01000016 ORZ36500.1 UZAN01040947 VDP71465.1 QUTC01005173 QUTF01005337 RHY59796.1 RHZ41238.1 MZMZ02001578 RQM28967.1 QUSZ01004019 QUTA01003360 QUTB01005544 QUTD01006158 QUTE01010019 RHY16237.1 RHY23908.1 RHY54772.1 RHY57150.1 RHZ15741.1 LYON01000019 OON11046.1 AEFK01219405 KZ150780 OUM67922.1 UYRU01000240 VDK29767.1 OON11044.1 JH817566 EKC36241.1 OON11045.1 MCFG01000009 ORX87413.1 BEYU01000008 GBG24782.1 MRZV01001920 PIK35361.1 MCOG01000109 ORY46164.1 KE560403 EPZ36886.1 CM004467 OCT96698.1 MBDO02000119 MBAD02000319 RLN62543.1 RLN69866.1 EFA00815.1 KQ976464 KYM84506.1 GL768066 EFZ12612.1 GG738859 EFC46234.1 GBEZ01023412 JAC63474.1 GBEZ01013889 JAC72139.1 ANIZ01002402 ETI40905.1 LNFO01005959 KUF75845.1 ANIY01002864 ETP38830.1 KI687601 KI681028 KI694266 ETK81016.1 ETL87708.1 ETM40940.1 LNFP01000197 KUF95892.1 ANIX01002729 ETP10677.1 MJFZ01000285 RAW32256.1 JH993003 EKX44720.1 CCKQ01015189 CDW86988.1 KI669564 ETN20797.1 KI674337 ETL34441.1 AMCR01020552 EJY66580.1 MCGO01000025 ORY43341.1 GL376632
NWSH01000682 PCG74817.1 KQ459232 KPJ02743.1 KQ461155 KPJ08195.1 JTDY01003165 KOB70028.1 DS235346 EEB15030.1 GEZM01075064 JAV64310.1 GEZM01075065 JAV64309.1 KZ430352 PIS84031.1 KV895655 OON17252.1 DF145157 GAA38980.2 KL596670 KER29860.1 NIRI01001908 RJW64567.1 UZAK01000104 VDO60005.1 GL882981 EGF75858.1 FNXT01001367 SZX79458.1 UZAL01027264 VDP30882.1 KQ965736 KXS20284.1 GL882890 EGF78030.1 DS469597 EDO40032.1 DS022311 OAJ44134.1 KQ977279 KYN04337.1 UZAI01000574 VDO54487.1 HAAD01003606 CDG69838.1 NOWV01000096 RDD40624.1 KL250496 KGB32247.1 KQ971322 KYB28600.1 OAJ44130.1 RCHS01001219 RMX54659.1 KQ982182 KYQ59225.1 HE601625 CCD77381.1 AAGJ04109380 AAGJ04109381 KQ971358 KYB25987.1 LSMT01000358 PFX19502.1 GEDC01008904 GEDC01008453 JAS28394.1 JAS28845.1 MCFH01000010 ORX54783.1 GEDC01025173 JAS12125.1 KQ980880 KYN11991.1 BDRX01000033 GBF92555.1 KQ981382 KYN42081.1 GEDC01010680 JAS26618.1 FN327264 CAX82988.1 AHAT01001233 FR907431 CDQ86682.1 MCFL01000016 ORZ36500.1 UZAN01040947 VDP71465.1 QUTC01005173 QUTF01005337 RHY59796.1 RHZ41238.1 MZMZ02001578 RQM28967.1 QUSZ01004019 QUTA01003360 QUTB01005544 QUTD01006158 QUTE01010019 RHY16237.1 RHY23908.1 RHY54772.1 RHY57150.1 RHZ15741.1 LYON01000019 OON11046.1 AEFK01219405 KZ150780 OUM67922.1 UYRU01000240 VDK29767.1 OON11044.1 JH817566 EKC36241.1 OON11045.1 MCFG01000009 ORX87413.1 BEYU01000008 GBG24782.1 MRZV01001920 PIK35361.1 MCOG01000109 ORY46164.1 KE560403 EPZ36886.1 CM004467 OCT96698.1 MBDO02000119 MBAD02000319 RLN62543.1 RLN69866.1 EFA00815.1 KQ976464 KYM84506.1 GL768066 EFZ12612.1 GG738859 EFC46234.1 GBEZ01023412 JAC63474.1 GBEZ01013889 JAC72139.1 ANIZ01002402 ETI40905.1 LNFO01005959 KUF75845.1 ANIY01002864 ETP38830.1 KI687601 KI681028 KI694266 ETK81016.1 ETL87708.1 ETM40940.1 LNFP01000197 KUF95892.1 ANIX01002729 ETP10677.1 MJFZ01000285 RAW32256.1 JH993003 EKX44720.1 CCKQ01015189 CDW86988.1 KI669564 ETN20797.1 KI674337 ETL34441.1 AMCR01020552 EJY66580.1 MCGO01000025 ORY43341.1 GL376632
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000053240
UP000037510
+ More
UP000009046 UP000050789 UP000007241 UP000256970 UP000050791 UP000070544 UP000001593 UP000077115 UP000078542 UP000050790 UP000277204 UP000253843 UP000007266 UP000275408 UP000075809 UP000008854 UP000007110 UP000050792 UP000225706 UP000193719 UP000078492 UP000247498 UP000078541 UP000018468 UP000193380 UP000193411 UP000050740 UP000189906 UP000007648 UP000195826 UP000005408 UP000193944 UP000230750 UP000002280 UP000265140 UP000193920 UP000030755 UP000186698 UP000277300 UP000284657 UP000007875 UP000078540 UP000006671 UP000018721 UP000052943 UP000018948 UP000053236 UP000054423 UP000054532 UP000018958 UP000251314 UP000011087 UP000018817 UP000053864 UP000193642
UP000009046 UP000050789 UP000007241 UP000256970 UP000050791 UP000070544 UP000001593 UP000077115 UP000078542 UP000050790 UP000277204 UP000253843 UP000007266 UP000275408 UP000075809 UP000008854 UP000007110 UP000050792 UP000225706 UP000193719 UP000078492 UP000247498 UP000078541 UP000018468 UP000193380 UP000193411 UP000050740 UP000189906 UP000007648 UP000195826 UP000005408 UP000193944 UP000230750 UP000002280 UP000265140 UP000193920 UP000030755 UP000186698 UP000277300 UP000284657 UP000007875 UP000078540 UP000006671 UP000018721 UP000052943 UP000018948 UP000053236 UP000054423 UP000054532 UP000018958 UP000251314 UP000011087 UP000018817 UP000053864 UP000193642
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JCN0
A0A2H1WY89
A0A212FAQ0
A0A2A4JRZ8
A0A194QB24
A0A194QT66
+ More
A0A0L7L3F1 E0VNS4 A0A1Y1KVV6 A0A1Y1KSC0 A0A2H1C9E3 A0A1S8WRZ2 H2KVV4 A0A075A2R1 A0A3R7FDP8 A0A183JBY3 F4PFY6 A0A383WPS1 A0A183NUU2 A0A139AUA1 F4PAA1 A7S8B1 A0A177WWW9 A0A195CUY8 A0A183LES3 T2MC62 A0A369S2I1 A0A094ZES1 A0A139WKX7 A0A177WXM0 A0A3M6UM28 A0A151XFY8 G4VBV7 A0A3Q0K433 A0A3Q0KCL4 A0A1W2W8S4 A0A3Q0K1H9 W4XZK1 A0A139WDP5 A0A183RSH0 A0A2B4RTF4 A0A1B6DRU8 A0A1Y1VFA7 A0A1B6CFK9 A0A151IVW0 A0A2V0P647 A0A195FNU1 A0A1B6DLU2 C7TZG2 W5NNC8 A0A060Y5A5 A0A1Y2HPI9 A0A183AAR5 A0A397DBX2 A0A3R7WKB3 A0A397D814 A0A1S8W9A1 G3VZN1 A0A1Y3NND5 A0A3P6P9G5 A0A1S8W9A2 K1RPB0 A0A1S8W9A8 A0A1Y1XNV4 A0A2R5G1A7 A0A2G8JHZ2 F6SBS0 A0A3P8Y293 A0A1Y2CH53 A0A075B3E6 A0A1L8HKU2 A0A3F2RRV2 D6WDQ7 H2ZR54 A0A195BJ87 E9J3L0 D2VA28 A0A061QYD0 A0A061RN40 V9ENT3 A0A0W8BVL1 W2YVK0 W2GFK7 A0A0W8DHT1 W2WK06 A0A329S8J5 L1J9H4 A0A078B1C9 W2R600 W2IJX0 J9I7B9 A0A1Y2C8H9 K3WVZ3
A0A0L7L3F1 E0VNS4 A0A1Y1KVV6 A0A1Y1KSC0 A0A2H1C9E3 A0A1S8WRZ2 H2KVV4 A0A075A2R1 A0A3R7FDP8 A0A183JBY3 F4PFY6 A0A383WPS1 A0A183NUU2 A0A139AUA1 F4PAA1 A7S8B1 A0A177WWW9 A0A195CUY8 A0A183LES3 T2MC62 A0A369S2I1 A0A094ZES1 A0A139WKX7 A0A177WXM0 A0A3M6UM28 A0A151XFY8 G4VBV7 A0A3Q0K433 A0A3Q0KCL4 A0A1W2W8S4 A0A3Q0K1H9 W4XZK1 A0A139WDP5 A0A183RSH0 A0A2B4RTF4 A0A1B6DRU8 A0A1Y1VFA7 A0A1B6CFK9 A0A151IVW0 A0A2V0P647 A0A195FNU1 A0A1B6DLU2 C7TZG2 W5NNC8 A0A060Y5A5 A0A1Y2HPI9 A0A183AAR5 A0A397DBX2 A0A3R7WKB3 A0A397D814 A0A1S8W9A1 G3VZN1 A0A1Y3NND5 A0A3P6P9G5 A0A1S8W9A2 K1RPB0 A0A1S8W9A8 A0A1Y1XNV4 A0A2R5G1A7 A0A2G8JHZ2 F6SBS0 A0A3P8Y293 A0A1Y2CH53 A0A075B3E6 A0A1L8HKU2 A0A3F2RRV2 D6WDQ7 H2ZR54 A0A195BJ87 E9J3L0 D2VA28 A0A061QYD0 A0A061RN40 V9ENT3 A0A0W8BVL1 W2YVK0 W2GFK7 A0A0W8DHT1 W2WK06 A0A329S8J5 L1J9H4 A0A078B1C9 W2R600 W2IJX0 J9I7B9 A0A1Y2C8H9 K3WVZ3
Ontologies
PANTHER
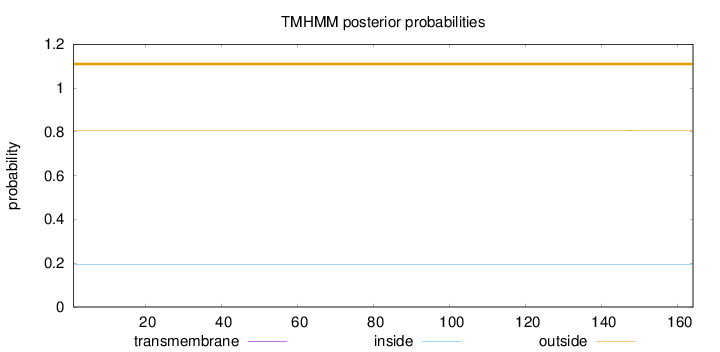
Topology
Length:
164
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00127
Exp number, first 60 AAs:
0
Total prob of N-in:
0.19454
outside
1 - 164
Population Genetic Test Statistics
Pi
322.579168
Theta
162.746133
Tajima's D
3.399029
CLR
0.570225
CSRT
0.992950352482376
Interpretation
Uncertain
