Gene
KWMTBOMO10257
Pre Gene Modal
BGIBMGA007281
Annotation
hypothetical_protein_RR46_09944_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.776
Sequence
CDS
ATGATACCACGATGGAGGAAAACTGTATCGAGATTGTTCCTAATTCCCCAGCGGTATGTCTTGGGCATCATGGGTCTGTTGGCTGTGTGCAACGCGTACACCATGCGAGTTTGCCTCAACCTGGCCGTCACTCAAATGGTTAATACAACAAAACACGAAGACGCGCATTTTGATCCTGACGCGTGTCCTGACGACAGCGAAATACTGACCAACGGAACAGTTAGCTTGAAACCGTACGCTATATTCGATTGGTCCGAATCAACGCAAGGCCTGATACTCAGCGGGTTTTACTATGGATACGCTATAACTCATGTCCCTGGAGGATACTTGGCGGAAAAATACGGAGGGAAATGGACTTTGGGAATAGGTCTTCTGAGTACAGCTATCTTCACGCTCATGACGCCCGTAGTTGTGAAAGCTGGGGGAGCTACTTGGCTATTCATTTTGAGAGTATTACAAGGAATGGGTGAGGGTCCAACAATGCCGGCTCTGATGATAGTACTGGCTCGATGGGTTCCACCCCATGAACGATCACGGCAAGGAGCTTTGGTCTTCGGTGGAGCACAAATAGGAAATATATTCGGTTCTTTCATGTCAGGATTCTTGATGGCTGGAGATGGTGATTGGGCTAATGTGTTTTATTTCTTCGGATGCTTTGGTGTACTATGGTTCGTGGCTTGGAGTATACTTTGCTTCAGTACACCAAACACTCACCCGTATATTTCTGAAGAGGAGTTGAATTATCTTAATGAAACAGTAACCAGTGCTGATACAAAAAGCGTCAGAGACCCCGTGCCTTGGAAAGCGATTGTCAGATCAGTGCCTGTTTGGGCATTATTGTTCGCTGCTGTTGGTCACGACTGGGGCTACTACACAATGGTGACAGATTTACCGAAATACATGACAGACGTTCTTAAATTCAATATCGCTACAACTGGAACGCTAACTGCGATTCCGTATCTCGCCATGTGGATAAGCTCATTTATATTTGGATGGGCCTGTGATTTTTGCGTGAAAAAGAATTGGCATTCCATTAAGACCGGCCGGATCATTCATACGACTATAGCTGCGACGGGGCCCGCAATATGTATCATATTGGCTTCATATGCGGGCTGTGACCGTTTTGCCGCCGTCGCCTATTTCATCGCATCTATGGCTCTCATGGGAGGATTCTACAGTGGAATGAAGGTGAATGCTTTAGACTTGGCACCGAATTACGCGGGTTCCTTGACCGCCATGATCAACGGGACGTCAACAATATCGGGAATCATCACGCCGTATCTAATTGGGCTATTAACGCCAGACTCAACCCTGGTACAGTGGCGTGTCGCGTTTTGGGTTTGTTTCGCCGTTCTGGTTGGAACGAACGTCGTGTACATAATTTGGGCTGACGGGAAACGTCAGTGGTGGGATGACGTGCGTCAATACGGTTATCCTGCTGATTGGAAACACGGTCCCCTGAAAATGGCGGACGAAGACAAAGAGAAAAGTAAAAAAGCCGAAGCTAATGATTCCGGGAAGTGA
Protein
MIPRWRKTVSRLFLIPQRYVLGIMGLLAVCNAYTMRVCLNLAVTQMVNTTKHEDAHFDPDACPDDSEILTNGTVSLKPYAIFDWSESTQGLILSGFYYGYAITHVPGGYLAEKYGGKWTLGIGLLSTAIFTLMTPVVVKAGGATWLFILRVLQGMGEGPTMPALMIVLARWVPPHERSRQGALVFGGAQIGNIFGSFMSGFLMAGDGDWANVFYFFGCFGVLWFVAWSILCFSTPNTHPYISEEELNYLNETVTSADTKSVRDPVPWKAIVRSVPVWALLFAAVGHDWGYYTMVTDLPKYMTDVLKFNIATTGTLTAIPYLAMWISSFIFGWACDFCVKKNWHSIKTGRIIHTTIAATGPAICIILASYAGCDRFAAVAYFIASMALMGGFYSGMKVNALDLAPNYAGSLTAMINGTSTISGIITPYLIGLLTPDSTLVQWRVAFWVCFAVLVGTNVVYIIWADGKRQWWDDVRQYGYPADWKHGPLKMADEDKEKSKKAEANDSGK
Summary
Uniprot
A0A2A4JNF2
H9JCN5
A0A194QH21
A0A3S2LXE2
A0A194QRT2
A0A2H1VXA1
+ More
A0A212FAP5 A0A2A4JS02 B2DBK3 A0A194QX98 A0A212FBW9 A0A194QRQ6 A0A3S2LGW2 H9JCN6 A0A194QBD2 A0A2A4JMR8 A0A2A4JNR0 H9JCN2 A0A212FBV8 A0A194QCN0 A0A194QS39 A0A3S2NNZ7 A0A194QB85 A0A2H1WRZ2 A0A2A4J376 H9JCM4 A0A212FEY9 V5GIW1 D6WY02 A0A1Y1KCZ6 A0A139WCX9 A0A2J7PXI4 A0A1Y1LE57 A0A0L7LVJ0 A0A0T6ASN5 A0A2M4BKX6 A0A2M3Z085 A0A2M4ARE2 A0A2M4AQL6 T1DQU9 A0A2M4BKU0 A0A2M4CV07 A0A1W4WUJ3 A0A182QUA2 A0A1S4GW12 A0A2C9H8P8 A0A2C9GS98 A0A067QQE2 Q17KJ1 A0A1L8DEM3 A0A182SZC7 A0A023EW05 A0A182J295 A0A023ET89 A0A1Q3EYV7 A0A3Q0J1H3 A0A1Q3EY35 A0A1Q3EYY5 A0A336K4P4 A0A084WHC7 A0A0K8VUW1 W8AKV5 A0A1S4GW29 A0A182L2Q9 A0A2C9H8Q5 A0A1S4EDR5 A0A1W4WT82 N6TY97 B4MZL1 B3MLH2 A0A1I8P1U7 A0A3B0K8S9 A0A182H8G9 A0A0A1XDB3 B4KJS0 A0A1Q3EY25 A0A1A9XFR6 B0WNU4 B4LQX0 T1IDQ4 A0A1A9Z8Q6 A0A182UMX2 Q29NJ2 B4GQI5 A0A0P4VSR8 A0A3Q0IWA8 A0A1W4UV06 A0A195DN90 A0A182XZT2 B4JQX8 A0A182F2R3 A0A3L8DRX0 Q7Q367 A0A0C9RCQ7 A0A232FJM4 N6TSG1 B4P1W9 A0A1I8NKQ2 U4TT62
A0A212FAP5 A0A2A4JS02 B2DBK3 A0A194QX98 A0A212FBW9 A0A194QRQ6 A0A3S2LGW2 H9JCN6 A0A194QBD2 A0A2A4JMR8 A0A2A4JNR0 H9JCN2 A0A212FBV8 A0A194QCN0 A0A194QS39 A0A3S2NNZ7 A0A194QB85 A0A2H1WRZ2 A0A2A4J376 H9JCM4 A0A212FEY9 V5GIW1 D6WY02 A0A1Y1KCZ6 A0A139WCX9 A0A2J7PXI4 A0A1Y1LE57 A0A0L7LVJ0 A0A0T6ASN5 A0A2M4BKX6 A0A2M3Z085 A0A2M4ARE2 A0A2M4AQL6 T1DQU9 A0A2M4BKU0 A0A2M4CV07 A0A1W4WUJ3 A0A182QUA2 A0A1S4GW12 A0A2C9H8P8 A0A2C9GS98 A0A067QQE2 Q17KJ1 A0A1L8DEM3 A0A182SZC7 A0A023EW05 A0A182J295 A0A023ET89 A0A1Q3EYV7 A0A3Q0J1H3 A0A1Q3EY35 A0A1Q3EYY5 A0A336K4P4 A0A084WHC7 A0A0K8VUW1 W8AKV5 A0A1S4GW29 A0A182L2Q9 A0A2C9H8Q5 A0A1S4EDR5 A0A1W4WT82 N6TY97 B4MZL1 B3MLH2 A0A1I8P1U7 A0A3B0K8S9 A0A182H8G9 A0A0A1XDB3 B4KJS0 A0A1Q3EY25 A0A1A9XFR6 B0WNU4 B4LQX0 T1IDQ4 A0A1A9Z8Q6 A0A182UMX2 Q29NJ2 B4GQI5 A0A0P4VSR8 A0A3Q0IWA8 A0A1W4UV06 A0A195DN90 A0A182XZT2 B4JQX8 A0A182F2R3 A0A3L8DRX0 Q7Q367 A0A0C9RCQ7 A0A232FJM4 N6TSG1 B4P1W9 A0A1I8NKQ2 U4TT62
Pubmed
EMBL
NWSH01000970
PCG73349.1
BABH01034602
KQ459232
KPJ02741.1
RSAL01000130
+ More
RVE46449.1 KQ461155 KPJ08197.1 ODYU01004960 SOQ45356.1 AGBW02009440 OWR50799.1 NWSH01000682 PCG74815.1 AB264687 BAG30762.1 KPJ02742.1 KPJ08196.1 AGBW02009266 OWR51223.1 KPJ08198.1 RVE46451.1 BABH01034604 KPJ02739.1 PCG73347.1 PCG73348.1 BABH01034596 OWR51222.1 KPJ02740.1 KPJ08134.1 RSAL01000189 RVE44729.1 KPJ02803.1 ODYU01010612 SOQ55840.1 NWSH01003662 PCG65964.1 BABH01034647 AGBW02008878 OWR52311.1 GALX01004487 JAB63979.1 KQ971362 EFA08924.2 GEZM01088379 JAV58231.1 KYB25747.1 NEVH01020858 PNF21048.1 GEZM01058093 JAV71913.1 JTDY01000017 KOB79399.1 LJIG01022894 KRT78184.1 GGFJ01004472 MBW53613.1 GGFM01001173 MBW21924.1 GGFK01010022 MBW43343.1 GGFK01009753 MBW43074.1 GAMD01000950 JAB00641.1 GGFJ01004473 MBW53614.1 GGFL01004975 MBW69153.1 AXCN02000700 AAAB01008964 APCN01005199 KK853134 KDR10840.1 CH477223 EAT47235.1 GFDF01009257 JAV04827.1 GAPW01001089 JAC12509.1 GAPW01001090 JAC12508.1 GFDL01014561 JAV20484.1 GFDL01014832 JAV20213.1 GFDL01014539 JAV20506.1 UFQS01000047 UFQT01000047 SSW98548.1 SSX18934.1 ATLV01023802 KE525346 KFB49621.1 GDHF01020879 GDHF01009648 JAI31435.1 JAI42666.1 GAMC01021217 GAMC01021215 JAB85338.1 APGK01057054 APGK01057055 APGK01057056 APGK01057057 APGK01057058 APGK01057059 KB741277 ENN71267.1 CH963920 EDW77796.1 CH902620 EDV31721.1 OUUW01000006 SPP81431.1 JXUM01029060 KQ560841 KXJ80654.1 GBXI01005361 JAD08931.1 CH933807 EDW11515.1 GFDL01014836 JAV20209.1 DS232015 EDS31939.1 CH940649 EDW64509.1 ACPB03000198 CH379059 EAL34227.2 CH479187 EDW39857.1 GDKW01002574 JAI54021.1 KQ980713 KYN14370.1 CH916372 EDV99308.1 QOIP01000005 RLU23056.1 EAA12474.3 GBYB01006080 JAG75847.1 NNAY01000124 OXU30740.1 ENN71266.1 CM000157 EDW88140.1 KB631617 ERL84714.1
RVE46449.1 KQ461155 KPJ08197.1 ODYU01004960 SOQ45356.1 AGBW02009440 OWR50799.1 NWSH01000682 PCG74815.1 AB264687 BAG30762.1 KPJ02742.1 KPJ08196.1 AGBW02009266 OWR51223.1 KPJ08198.1 RVE46451.1 BABH01034604 KPJ02739.1 PCG73347.1 PCG73348.1 BABH01034596 OWR51222.1 KPJ02740.1 KPJ08134.1 RSAL01000189 RVE44729.1 KPJ02803.1 ODYU01010612 SOQ55840.1 NWSH01003662 PCG65964.1 BABH01034647 AGBW02008878 OWR52311.1 GALX01004487 JAB63979.1 KQ971362 EFA08924.2 GEZM01088379 JAV58231.1 KYB25747.1 NEVH01020858 PNF21048.1 GEZM01058093 JAV71913.1 JTDY01000017 KOB79399.1 LJIG01022894 KRT78184.1 GGFJ01004472 MBW53613.1 GGFM01001173 MBW21924.1 GGFK01010022 MBW43343.1 GGFK01009753 MBW43074.1 GAMD01000950 JAB00641.1 GGFJ01004473 MBW53614.1 GGFL01004975 MBW69153.1 AXCN02000700 AAAB01008964 APCN01005199 KK853134 KDR10840.1 CH477223 EAT47235.1 GFDF01009257 JAV04827.1 GAPW01001089 JAC12509.1 GAPW01001090 JAC12508.1 GFDL01014561 JAV20484.1 GFDL01014832 JAV20213.1 GFDL01014539 JAV20506.1 UFQS01000047 UFQT01000047 SSW98548.1 SSX18934.1 ATLV01023802 KE525346 KFB49621.1 GDHF01020879 GDHF01009648 JAI31435.1 JAI42666.1 GAMC01021217 GAMC01021215 JAB85338.1 APGK01057054 APGK01057055 APGK01057056 APGK01057057 APGK01057058 APGK01057059 KB741277 ENN71267.1 CH963920 EDW77796.1 CH902620 EDV31721.1 OUUW01000006 SPP81431.1 JXUM01029060 KQ560841 KXJ80654.1 GBXI01005361 JAD08931.1 CH933807 EDW11515.1 GFDL01014836 JAV20209.1 DS232015 EDS31939.1 CH940649 EDW64509.1 ACPB03000198 CH379059 EAL34227.2 CH479187 EDW39857.1 GDKW01002574 JAI54021.1 KQ980713 KYN14370.1 CH916372 EDV99308.1 QOIP01000005 RLU23056.1 EAA12474.3 GBYB01006080 JAG75847.1 NNAY01000124 OXU30740.1 ENN71266.1 CM000157 EDW88140.1 KB631617 ERL84714.1
Proteomes
UP000218220
UP000005204
UP000053268
UP000283053
UP000053240
UP000007151
+ More
UP000007266 UP000235965 UP000037510 UP000192223 UP000075886 UP000076407 UP000075840 UP000027135 UP000008820 UP000075901 UP000075880 UP000079169 UP000030765 UP000075882 UP000019118 UP000007798 UP000007801 UP000095300 UP000268350 UP000069940 UP000249989 UP000009192 UP000092443 UP000002320 UP000008792 UP000015103 UP000092445 UP000075903 UP000001819 UP000008744 UP000192221 UP000078492 UP000076408 UP000001070 UP000069272 UP000279307 UP000007062 UP000215335 UP000002282 UP000095301 UP000030742
UP000007266 UP000235965 UP000037510 UP000192223 UP000075886 UP000076407 UP000075840 UP000027135 UP000008820 UP000075901 UP000075880 UP000079169 UP000030765 UP000075882 UP000019118 UP000007798 UP000007801 UP000095300 UP000268350 UP000069940 UP000249989 UP000009192 UP000092443 UP000002320 UP000008792 UP000015103 UP000092445 UP000075903 UP000001819 UP000008744 UP000192221 UP000078492 UP000076408 UP000001070 UP000069272 UP000279307 UP000007062 UP000215335 UP000002282 UP000095301 UP000030742
Pfam
PF07690 MFS_1
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A2A4JNF2
H9JCN5
A0A194QH21
A0A3S2LXE2
A0A194QRT2
A0A2H1VXA1
+ More
A0A212FAP5 A0A2A4JS02 B2DBK3 A0A194QX98 A0A212FBW9 A0A194QRQ6 A0A3S2LGW2 H9JCN6 A0A194QBD2 A0A2A4JMR8 A0A2A4JNR0 H9JCN2 A0A212FBV8 A0A194QCN0 A0A194QS39 A0A3S2NNZ7 A0A194QB85 A0A2H1WRZ2 A0A2A4J376 H9JCM4 A0A212FEY9 V5GIW1 D6WY02 A0A1Y1KCZ6 A0A139WCX9 A0A2J7PXI4 A0A1Y1LE57 A0A0L7LVJ0 A0A0T6ASN5 A0A2M4BKX6 A0A2M3Z085 A0A2M4ARE2 A0A2M4AQL6 T1DQU9 A0A2M4BKU0 A0A2M4CV07 A0A1W4WUJ3 A0A182QUA2 A0A1S4GW12 A0A2C9H8P8 A0A2C9GS98 A0A067QQE2 Q17KJ1 A0A1L8DEM3 A0A182SZC7 A0A023EW05 A0A182J295 A0A023ET89 A0A1Q3EYV7 A0A3Q0J1H3 A0A1Q3EY35 A0A1Q3EYY5 A0A336K4P4 A0A084WHC7 A0A0K8VUW1 W8AKV5 A0A1S4GW29 A0A182L2Q9 A0A2C9H8Q5 A0A1S4EDR5 A0A1W4WT82 N6TY97 B4MZL1 B3MLH2 A0A1I8P1U7 A0A3B0K8S9 A0A182H8G9 A0A0A1XDB3 B4KJS0 A0A1Q3EY25 A0A1A9XFR6 B0WNU4 B4LQX0 T1IDQ4 A0A1A9Z8Q6 A0A182UMX2 Q29NJ2 B4GQI5 A0A0P4VSR8 A0A3Q0IWA8 A0A1W4UV06 A0A195DN90 A0A182XZT2 B4JQX8 A0A182F2R3 A0A3L8DRX0 Q7Q367 A0A0C9RCQ7 A0A232FJM4 N6TSG1 B4P1W9 A0A1I8NKQ2 U4TT62
A0A212FAP5 A0A2A4JS02 B2DBK3 A0A194QX98 A0A212FBW9 A0A194QRQ6 A0A3S2LGW2 H9JCN6 A0A194QBD2 A0A2A4JMR8 A0A2A4JNR0 H9JCN2 A0A212FBV8 A0A194QCN0 A0A194QS39 A0A3S2NNZ7 A0A194QB85 A0A2H1WRZ2 A0A2A4J376 H9JCM4 A0A212FEY9 V5GIW1 D6WY02 A0A1Y1KCZ6 A0A139WCX9 A0A2J7PXI4 A0A1Y1LE57 A0A0L7LVJ0 A0A0T6ASN5 A0A2M4BKX6 A0A2M3Z085 A0A2M4ARE2 A0A2M4AQL6 T1DQU9 A0A2M4BKU0 A0A2M4CV07 A0A1W4WUJ3 A0A182QUA2 A0A1S4GW12 A0A2C9H8P8 A0A2C9GS98 A0A067QQE2 Q17KJ1 A0A1L8DEM3 A0A182SZC7 A0A023EW05 A0A182J295 A0A023ET89 A0A1Q3EYV7 A0A3Q0J1H3 A0A1Q3EY35 A0A1Q3EYY5 A0A336K4P4 A0A084WHC7 A0A0K8VUW1 W8AKV5 A0A1S4GW29 A0A182L2Q9 A0A2C9H8Q5 A0A1S4EDR5 A0A1W4WT82 N6TY97 B4MZL1 B3MLH2 A0A1I8P1U7 A0A3B0K8S9 A0A182H8G9 A0A0A1XDB3 B4KJS0 A0A1Q3EY25 A0A1A9XFR6 B0WNU4 B4LQX0 T1IDQ4 A0A1A9Z8Q6 A0A182UMX2 Q29NJ2 B4GQI5 A0A0P4VSR8 A0A3Q0IWA8 A0A1W4UV06 A0A195DN90 A0A182XZT2 B4JQX8 A0A182F2R3 A0A3L8DRX0 Q7Q367 A0A0C9RCQ7 A0A232FJM4 N6TSG1 B4P1W9 A0A1I8NKQ2 U4TT62
PDB
6E9N
E-value=6.31807e-15,
Score=198
Ontologies
GO
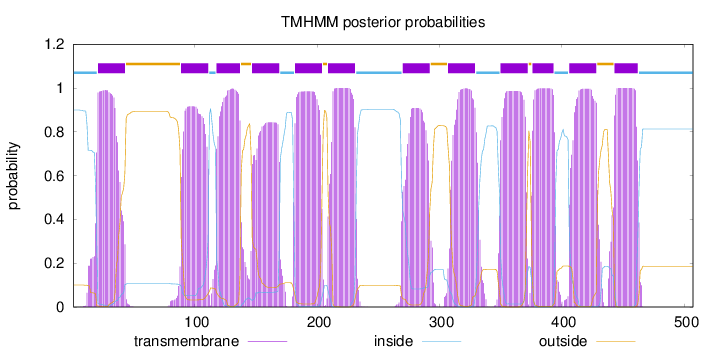
Topology
Length:
507
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
252.98874
Exp number, first 60 AAs:
21.42269
Total prob of N-in:
0.89928
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 88
TMhelix
89 - 111
inside
112 - 117
TMhelix
118 - 137
outside
138 - 146
TMhelix
147 - 169
inside
170 - 181
TMhelix
182 - 204
outside
205 - 208
TMhelix
209 - 231
inside
232 - 269
TMhelix
270 - 292
outside
293 - 306
TMhelix
307 - 329
inside
330 - 349
TMhelix
350 - 372
outside
373 - 375
TMhelix
376 - 393
inside
394 - 405
TMhelix
406 - 428
outside
429 - 442
TMhelix
443 - 462
inside
463 - 507
Population Genetic Test Statistics
Pi
171.406642
Theta
148.996561
Tajima's D
-1.323303
CLR
0.52635
CSRT
0.0854957252137393
Interpretation
Uncertain
