Gene
KWMTBOMO10248 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007311
Annotation
40S_ribosomal_protein_SA_[Bombyx_mori]
Full name
40S ribosomal protein SA
Alternative Name
37 kDa laminin receptor precursor
37/67 kDa laminin receptor
67 kDa laminin receptor
Laminin receptor 1
Laminin-binding protein precursor p40
37/67 kDa laminin receptor
67 kDa laminin receptor
Laminin receptor 1
Laminin-binding protein precursor p40
Location in the cell
Cytoplasmic Reliability : 2.911
Sequence
CDS
ATGTCGGGAGGATTAGACGTGCTCGCCCTCAACGAGGAAGATGTCACCAAAATGCTTGCTGCAACCACCCATCTTGGGGCAGAAAATGTTAACTTCCAGATGGAGACCTATGTCTACAAACGACGTGCTGATGGTACCCATGTGATCAACTTGCGTCGTACCTGGGAAAAACTTGTTCTGGCTGCTCGTGCTGTCGTAGCCATCGAGAACCCCGCTGATGTGTTCGTCATCTCATCACGGCCCTTCGGTCAGCGTGCTGTACTGAAGTTTGCCGCGCACACCGGTGCTACGCCTATTGCGGGACGTTTCACACCAGGTGCTTTTACTAACCAGATCCAAGCTGCATTCCGTGAACCTCGTCTCTTGATTGTATTGGACCCTGCACAAGACCATCAACCCATTACTGAAGCTTCATATGTCAACATTCCTGTGATTGCTTTGTGCAACACAGACTCCCCACTAAGATTTGTGGACATTGCTATCCCATGCAACACCAAGTCTTCCCACTCTATTGGTTTGATGTGGTGGTTGTTGGCACGTGAAGTGCTGAGGCTTCGTGGTGTGCTTCCCCGTGACCAGCGCTGGGATGTTGTGGTTGATTTGTTCTTCTACCGTGACCCTGAAGAAAGTGAAAAGGATGAACAACAAGCCAAGGAACAGGCTGTGGTACCAGCTAAACCAGAAGTAGTTGCTCCTGTCCATGAAGACTGGAATGAAACACTGGAGCCAGTGGCTTCATGGGCTGAAGATACCGCCCCGCCTGCTGCACCCGCTGCAGCTCCTGCTTTTGGAGCACCCCCTGCTCAAGAAGAATGGTCTGCCCAGGTACAAGATGAGTGGAGCACAACTGCCGCCGCACCAGCCGCCGCCACCGCAGCGCCCTCGTGGGGTGGATCCGCTCAAGACTGGAGTGCTGCATAA
Protein
MSGGLDVLALNEEDVTKMLAATTHLGAENVNFQMETYVYKRRADGTHVINLRRTWEKLVLAARAVVAIENPADVFVISSRPFGQRAVLKFAAHTGATPIAGRFTPGAFTNQIQAAFREPRLLIVLDPAQDHQPITEASYVNIPVIALCNTDSPLRFVDIAIPCNTKSSHSIGLMWWLLAREVLRLRGVLPRDQRWDVVVDLFFYRDPEESEKDEQQAKEQAVVPAKPEVVAPVHEDWNETLEPVASWAEDTAPPAAPAAAPAFGAPPAQEEWSAQVQDEWSTTAAAPAAATAAPSWGGSAQDWSAA
Summary
Description
Required for the assembly and/or stability of the 40S ribosomal subunit. Required for the processing of the 20S rRNA-precursor to mature 18S rRNA in a late step of the maturation of 40S ribosomal subunits.
Required for the assembly and/or stability of the 40S ribosomal subunit. Required for the processing of the 20S rRNA-precursor to mature 18S rRNA in a late step of the maturation of 40S ribosomal subunits. Also functions as a cell surface receptor for laminin. Plays a role in cell adhesion to the basement membrane and in the consequent activation of signaling transduction pathways. May play a role in cell fate determination and tissue morphogenesis.
Required for the assembly and/or stability of the 40S ribosomal subunit. Required for the processing of the 20S rRNA-precursor to mature 18S rRNA in a late step of the maturation of 40S ribosomal subunits. Also functions as a cell surface receptor for laminin. Plays a role in cell adhesion to the basement membrane and in the consequent activation of signaling transduction pathways. May play a role in cell fate determination and tissue morphogenesis.
Subunit
Component of the small ribosomal subunit. Mature ribosomes consist of a small (40S) and a large (60S) subunit. The 40S subunit contains about 33 different proteins and 1 molecule of RNA (18S). The 60S subunit contains about 49 different proteins and 3 molecules of RNA (28S, 5.8S and 5S). Interacts with ribosomal protein S21.
Monomer (37LRP) and homodimer (67LR). Component of the small ribosomal subunit. Mature ribosomes consist of a small (40S) and a large (60S) subunit. The 40S subunit contains about 33 different proteins and 1 molecule of RNA (18S). The 60S subunit contains about 49 different proteins and 3 molecules of RNA (28S, 5.8S and 5S). Interacts with RPS21. Interacts with several laminins including at least LAMB1. Interacts with MDK.
Monomer (37LRP) and homodimer (67LR). Component of the small ribosomal subunit. Mature ribosomes consist of a small (40S) and a large (60S) subunit. The 40S subunit contains about 33 different proteins and 1 molecule of RNA (18S). The 60S subunit contains about 49 different proteins and 3 molecules of RNA (28S, 5.8S and 5S). Interacts with RPS21. Interacts with several laminins including at least LAMB1. Interacts with MDK.
Miscellaneous
This protein appears to have acquired a second function as a laminin receptor specifically in the vertebrate lineage.
It is thought that in vertebrates 37/67 kDa laminin receptor acquired a dual function during evolution. It developed from the ribosomal protein SA, playing an essential role in the protein biosynthesis lacking any laminin binding activity, to a cell surface receptor with laminin binding activity.
It is thought that in vertebrates 37/67 kDa laminin receptor acquired a dual function during evolution. It developed from the ribosomal protein SA, playing an essential role in the protein biosynthesis lacking any laminin binding activity, to a cell surface receptor with laminin binding activity.
Similarity
Belongs to the universal ribosomal protein uS2 family.
Keywords
Complete proteome
Cytoplasm
Reference proteome
Ribonucleoprotein
Ribosomal protein
Acetylation
Cell membrane
Membrane
Nucleus
Receptor
Repeat
Feature
chain 40S ribosomal protein SA
Uniprot
H9JCR4
Q5UAP4
A0A3G1T1D1
A0A194QB13
A0A194QRT7
I4DM16
+ More
A0A3S2NXV9 E7DZ30 A0A2H1WG78 A0A2A4JYM1 S4PWY4 I4DNB4 G0ZEE8 A0A212FIS8 A0A0L7LWA1 A0A2J7RGF5 A0A067R227 A0A1W4WZL7 Q0PXX8 A0A1B6CY39 A0A1B6LW65 E0VN22 A0A1J1IRJ4 A0A023F8Y8 A0A161TJI5 A0A0C9PRY9 U5EZJ0 E2BW62 A2I3Z2 E7D1C2 A0A1Y1KZ33 A0A1W2WBX0 H2XTB0 A0A0X7YDQ6 A0A093T4B5 A0A0P4VRM4 R4G8Q1 A0A069DS42 A0A0V0G2J1 A0A091JN11 H2Z279 A0A224XTF0 A0A091HWW7 A0A091HPQ8 A0A099ZLP8 A0A3L8SZB8 A0A093Q9Q3 A0A218UPN6 A0A093GUL4 A0A093C052 A0A0A0A076 A0A091QAI8 A0A091S358 A0A093EMY8 A0A091KW03 A0A094L9N9 A0A091EQM2 A0A093I9Y2 A0A091RCB6 A0A091SMP9 A0A087QH29 R7VPS0 D5XMH1 B5FXT6 G1NGU2 A0A226MJ30 A0A091FRP5 A0A091V0M7 A0A226PU73 R0JZY0 A0A1U7SHM5 P50890 M7B099 A0A3Q0FWN0 A0A091PBH2 K4G2X5 A0A093DK92 U3J7Y9
A0A3S2NXV9 E7DZ30 A0A2H1WG78 A0A2A4JYM1 S4PWY4 I4DNB4 G0ZEE8 A0A212FIS8 A0A0L7LWA1 A0A2J7RGF5 A0A067R227 A0A1W4WZL7 Q0PXX8 A0A1B6CY39 A0A1B6LW65 E0VN22 A0A1J1IRJ4 A0A023F8Y8 A0A161TJI5 A0A0C9PRY9 U5EZJ0 E2BW62 A2I3Z2 E7D1C2 A0A1Y1KZ33 A0A1W2WBX0 H2XTB0 A0A0X7YDQ6 A0A093T4B5 A0A0P4VRM4 R4G8Q1 A0A069DS42 A0A0V0G2J1 A0A091JN11 H2Z279 A0A224XTF0 A0A091HWW7 A0A091HPQ8 A0A099ZLP8 A0A3L8SZB8 A0A093Q9Q3 A0A218UPN6 A0A093GUL4 A0A093C052 A0A0A0A076 A0A091QAI8 A0A091S358 A0A093EMY8 A0A091KW03 A0A094L9N9 A0A091EQM2 A0A093I9Y2 A0A091RCB6 A0A091SMP9 A0A087QH29 R7VPS0 D5XMH1 B5FXT6 G1NGU2 A0A226MJ30 A0A091FRP5 A0A091V0M7 A0A226PU73 R0JZY0 A0A1U7SHM5 P50890 M7B099 A0A3Q0FWN0 A0A091PBH2 K4G2X5 A0A093DK92 U3J7Y9
Pubmed
EMBL
BABH01041208
AB062685
AY769314
MG846933
AXY94785.1
KQ459232
+ More
KPJ02733.1 KQ461155 KPJ08202.1 AK402334 BAM18956.1 RSAL01000029 RVE51781.1 HQ424733 ADT80677.1 ODYU01008453 SOQ52078.1 NWSH01000429 PCG76472.1 GAIX01005983 JAA86577.1 AK402822 BAM19404.1 JF265026 AEL28857.1 AGBW02008335 OWR53636.1 JTDY01000012 KOB79436.1 NEVH01003784 PNF39921.1 KK853076 KDR11727.1 DQ673408 GEDC01018941 JAS18357.1 GEBQ01012058 JAT27919.1 DS235332 EEB14788.1 CVRI01000056 CRL01702.1 GBBI01000802 JAC17910.1 GEMB01000119 JAS02992.1 GBYB01004138 JAG73905.1 GANO01000709 JAB59162.1 GL451091 EFN80094.1 EF070495 HQ005870 ADV40166.1 GEZM01069071 JAV66662.1 EAAA01000818 KC507298 AHB12421.1 KL423107 KFW89494.1 GDKW01001528 JAI55067.1 GAHY01000582 JAA76928.1 GBGD01002343 JAC86546.1 GECL01003770 JAP02354.1 KK502279 KFP21952.1 GFTR01004716 JAW11710.1 KL217907 KFP00392.1 KL520075 KFO89308.1 KL894487 KGL81725.1 QUSF01000003 RLW11679.1 KL672330 KFW85678.1 MUZQ01000204 OWK55380.1 KL216618 KFV70752.1 KL236730 KFV09350.1 KL870344 KGL87949.1 KK690823 KFQ16571.1 KK941080 KFQ50699.1 KK373306 KFV44653.1 KK756306 KFP43450.1 KL353928 KFZ60774.1 KK718893 KFO59326.1 KL206987 KFV87932.1 KK812868 KFQ36857.1 KK478086 KFQ59643.1 KL225582 KFM00533.1 KB375329 AKCR02000094 EMC80727.1 PKK20975.1 DP001180 DP001179 ADH10545.1 DQ213582 DQ213584 DQ213585 MCFN01000769 OXB55305.1 KL447288 KFO71809.1 KL410402 KFQ96486.1 AWGT02000015 OXB83238.1 KB742877 EOB03226.1 X94368 KB589327 EMP25493.1 KK847598 KFP88875.1 JX207230 AFM85544.1 KN126788 KFU92660.1 ADON01054041
KPJ02733.1 KQ461155 KPJ08202.1 AK402334 BAM18956.1 RSAL01000029 RVE51781.1 HQ424733 ADT80677.1 ODYU01008453 SOQ52078.1 NWSH01000429 PCG76472.1 GAIX01005983 JAA86577.1 AK402822 BAM19404.1 JF265026 AEL28857.1 AGBW02008335 OWR53636.1 JTDY01000012 KOB79436.1 NEVH01003784 PNF39921.1 KK853076 KDR11727.1 DQ673408 GEDC01018941 JAS18357.1 GEBQ01012058 JAT27919.1 DS235332 EEB14788.1 CVRI01000056 CRL01702.1 GBBI01000802 JAC17910.1 GEMB01000119 JAS02992.1 GBYB01004138 JAG73905.1 GANO01000709 JAB59162.1 GL451091 EFN80094.1 EF070495 HQ005870 ADV40166.1 GEZM01069071 JAV66662.1 EAAA01000818 KC507298 AHB12421.1 KL423107 KFW89494.1 GDKW01001528 JAI55067.1 GAHY01000582 JAA76928.1 GBGD01002343 JAC86546.1 GECL01003770 JAP02354.1 KK502279 KFP21952.1 GFTR01004716 JAW11710.1 KL217907 KFP00392.1 KL520075 KFO89308.1 KL894487 KGL81725.1 QUSF01000003 RLW11679.1 KL672330 KFW85678.1 MUZQ01000204 OWK55380.1 KL216618 KFV70752.1 KL236730 KFV09350.1 KL870344 KGL87949.1 KK690823 KFQ16571.1 KK941080 KFQ50699.1 KK373306 KFV44653.1 KK756306 KFP43450.1 KL353928 KFZ60774.1 KK718893 KFO59326.1 KL206987 KFV87932.1 KK812868 KFQ36857.1 KK478086 KFQ59643.1 KL225582 KFM00533.1 KB375329 AKCR02000094 EMC80727.1 PKK20975.1 DP001180 DP001179 ADH10545.1 DQ213582 DQ213584 DQ213585 MCFN01000769 OXB55305.1 KL447288 KFO71809.1 KL410402 KFQ96486.1 AWGT02000015 OXB83238.1 KB742877 EOB03226.1 X94368 KB589327 EMP25493.1 KK847598 KFP88875.1 JX207230 AFM85544.1 KN126788 KFU92660.1 ADON01054041
Proteomes
UP000005204
UP000053268
UP000053240
UP000283053
UP000218220
UP000007151
+ More
UP000037510 UP000235965 UP000027135 UP000192223 UP000079169 UP000009046 UP000183832 UP000008237 UP000008144 UP000053119 UP000007875 UP000054308 UP000053641 UP000276834 UP000053258 UP000197619 UP000053875 UP000053858 UP000052976 UP000053584 UP000053286 UP000053872 UP000007754 UP000001645 UP000198323 UP000053760 UP000053283 UP000198419 UP000189705 UP000000539 UP000031443 UP000016666
UP000037510 UP000235965 UP000027135 UP000192223 UP000079169 UP000009046 UP000183832 UP000008237 UP000008144 UP000053119 UP000007875 UP000054308 UP000053641 UP000276834 UP000053258 UP000197619 UP000053875 UP000053858 UP000052976 UP000053584 UP000053286 UP000053872 UP000007754 UP000001645 UP000198323 UP000053760 UP000053283 UP000198419 UP000189705 UP000000539 UP000031443 UP000016666
Interpro
SUPFAM
SSF52313
SSF52313
CDD
ProteinModelPortal
H9JCR4
Q5UAP4
A0A3G1T1D1
A0A194QB13
A0A194QRT7
I4DM16
+ More
A0A3S2NXV9 E7DZ30 A0A2H1WG78 A0A2A4JYM1 S4PWY4 I4DNB4 G0ZEE8 A0A212FIS8 A0A0L7LWA1 A0A2J7RGF5 A0A067R227 A0A1W4WZL7 Q0PXX8 A0A1B6CY39 A0A1B6LW65 E0VN22 A0A1J1IRJ4 A0A023F8Y8 A0A161TJI5 A0A0C9PRY9 U5EZJ0 E2BW62 A2I3Z2 E7D1C2 A0A1Y1KZ33 A0A1W2WBX0 H2XTB0 A0A0X7YDQ6 A0A093T4B5 A0A0P4VRM4 R4G8Q1 A0A069DS42 A0A0V0G2J1 A0A091JN11 H2Z279 A0A224XTF0 A0A091HWW7 A0A091HPQ8 A0A099ZLP8 A0A3L8SZB8 A0A093Q9Q3 A0A218UPN6 A0A093GUL4 A0A093C052 A0A0A0A076 A0A091QAI8 A0A091S358 A0A093EMY8 A0A091KW03 A0A094L9N9 A0A091EQM2 A0A093I9Y2 A0A091RCB6 A0A091SMP9 A0A087QH29 R7VPS0 D5XMH1 B5FXT6 G1NGU2 A0A226MJ30 A0A091FRP5 A0A091V0M7 A0A226PU73 R0JZY0 A0A1U7SHM5 P50890 M7B099 A0A3Q0FWN0 A0A091PBH2 K4G2X5 A0A093DK92 U3J7Y9
A0A3S2NXV9 E7DZ30 A0A2H1WG78 A0A2A4JYM1 S4PWY4 I4DNB4 G0ZEE8 A0A212FIS8 A0A0L7LWA1 A0A2J7RGF5 A0A067R227 A0A1W4WZL7 Q0PXX8 A0A1B6CY39 A0A1B6LW65 E0VN22 A0A1J1IRJ4 A0A023F8Y8 A0A161TJI5 A0A0C9PRY9 U5EZJ0 E2BW62 A2I3Z2 E7D1C2 A0A1Y1KZ33 A0A1W2WBX0 H2XTB0 A0A0X7YDQ6 A0A093T4B5 A0A0P4VRM4 R4G8Q1 A0A069DS42 A0A0V0G2J1 A0A091JN11 H2Z279 A0A224XTF0 A0A091HWW7 A0A091HPQ8 A0A099ZLP8 A0A3L8SZB8 A0A093Q9Q3 A0A218UPN6 A0A093GUL4 A0A093C052 A0A0A0A076 A0A091QAI8 A0A091S358 A0A093EMY8 A0A091KW03 A0A094L9N9 A0A091EQM2 A0A093I9Y2 A0A091RCB6 A0A091SMP9 A0A087QH29 R7VPS0 D5XMH1 B5FXT6 G1NGU2 A0A226MJ30 A0A091FRP5 A0A091V0M7 A0A226PU73 R0JZY0 A0A1U7SHM5 P50890 M7B099 A0A3Q0FWN0 A0A091PBH2 K4G2X5 A0A093DK92 U3J7Y9
PDB
3BCH
E-value=3.79609e-97,
Score=904
Ontologies
GO
PANTHER
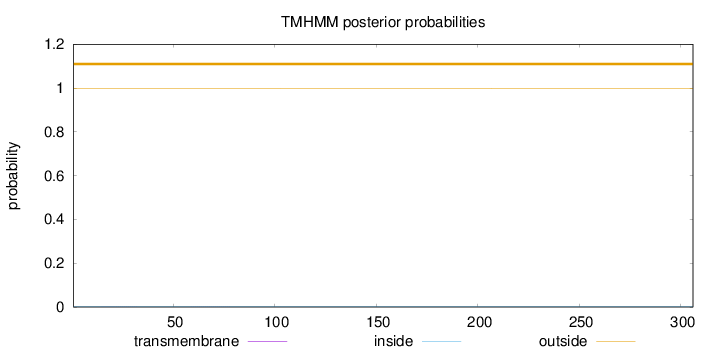
Topology
Subcellular location
Cytoplasm
Cell membrane 67LR is found at the surface of the plasma membrane, with its C-terminal laminin-binding domain accessible to extracellular ligands. 37LRP is found at the cell surface, in the cytoplasm and in the nucleus. With evidence from 1 publications.
Nucleus 67LR is found at the surface of the plasma membrane, with its C-terminal laminin-binding domain accessible to extracellular ligands. 37LRP is found at the cell surface, in the cytoplasm and in the nucleus. With evidence from 1 publications.
Cell membrane 67LR is found at the surface of the plasma membrane, with its C-terminal laminin-binding domain accessible to extracellular ligands. 37LRP is found at the cell surface, in the cytoplasm and in the nucleus. With evidence from 1 publications.
Nucleus 67LR is found at the surface of the plasma membrane, with its C-terminal laminin-binding domain accessible to extracellular ligands. 37LRP is found at the cell surface, in the cytoplasm and in the nucleus. With evidence from 1 publications.
Length:
306
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00974999999999999
Exp number, first 60 AAs:
0.00293
Total prob of N-in:
0.00268
outside
1 - 306
Population Genetic Test Statistics
Pi
220.260037
Theta
184.192878
Tajima's D
0.666065
CLR
0.683246
CSRT
0.55897205139743
Interpretation
Uncertain
