Gene
KWMTBOMO10247 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007310
Annotation
DRK_protein_[Bombyx_mori]
Full name
Protein E(sev)2B
+ More
Protein enhancer of sevenless 2B
Protein enhancer of sevenless 2B
Alternative Name
Downstream of receptor kinase
Protein enhancer of sevenless 2B
SH2-SH3 adapter protein drk
Protein enhancer of sevenless 2B
SH2-SH3 adapter protein drk
Location in the cell
Cytoplasmic Reliability : 2.653
Sequence
CDS
ATGGAGGCTCTAGCAAAACACGATTTTACTGCAACCGCAGATGATGAGCTCAGTTTTAGAAAAAATCAAGTATTAAAAATTCTAAACATGGAGGACGATATGAACTGGTACAGAGCTGAATTAGACGGGAAAGAAGGGCTCATTCCTAGTAATTACATACAAATGAAAAATCACAGTTGGTATTATGGGCGTATTACAAGAGCAGATGCAGAAAAACTCTTAGCAAACAAACCTGAAGGTGGCTTCTTGATAAGGATCAGCGAATCAAGCCCTGGCGACTTCTCCTTATCAGTCAAATGTCCGGACGGTGTCCAGCACTTCAAAGTATTAAGAGACGCCTCGAGCAAGTTCTTCCTTTGGGTCGTCAAGTTCAATTCGCTCAACGAACTAGTTGATTATCATCGTACCGCGTCTGTCAGTCGCCTTCAAGACGTTAAACTGCGCGACGTCGTCCCCGAAGAAATGTTGGTGCAGGCTCTATACGACTTCACCCCACAAGAGGCTGGCGAGTTGGAGTTTAGACGCGGTGACGTCATAACCGTGACAGATCGCTCCGACCAGCACTGGTGGCAGGTAAAGTGA
Protein
MEALAKHDFTATADDELSFRKNQVLKILNMEDDMNWYRAELDGKEGLIPSNYIQMKNHSWYYGRITRADAEKLLANKPEGGFLIRISESSPGDFSLSVKCPDGVQHFKVLRDASSKFFLWVVKFNSLNELVDYHRTASVSRLQDVKLRDVVPEEMLVQALYDFTPQEAGELEFRRGDVITVTDRSDQHWWQVK
Summary
Description
Required for proper signaling by sevenless. May act to stimulate the ability of Sos to catalyze Ras1 activation by linking sevenless and Sos in a signaling complex (By similarity).
Required for proper signaling by sevenless. May act to stimulate the ability of Sos to catalyze Ras1 activation by linking sevenless and Sos in a signaling complex.
Required for proper signaling by sevenless. May act to stimulate the ability of Sos to catalyze Ras1 activation by linking sevenless and Sos in a signaling complex.
Subunit
Interacts with autophosphorylated sev via SH2 domain and Sos and Dab via SH3 domains. Binds to tyrosine phosphorylated Dab via the SH2 domain (By similarity).
Interacts with autophosphorylated sev via SH2 domain and Sos, Dos and Dab via SH3 domains. Binds to tyrosine phosphorylated Dab via the SH2 domain.
Interacts with autophosphorylated sev via SH2 domain and Sos, Dos and Dab via SH3 domains. Binds to tyrosine phosphorylated Dab via the SH2 domain.
Similarity
Belongs to the GRB2/sem-5/DRK family.
Keywords
Membrane
Repeat
SH2 domain
SH3 domain
Transducer
3D-structure
Complete proteome
Reference proteome
Feature
chain Protein E(sev)2B
Uniprot
F2XHI6
A0A194QRR0
A0A194QB94
S4PH84
A0A2H1WG69
A0A1E1WLK8
+ More
A0A0L7LVH6 A0A212FIT9 A0A0K8TSK2 A0A2P8ZGQ7 A0A1B6E815 A0A1I8M7H0 A0A1S3D1F1 D3TPP1 A0A0L0C544 A0A146M7N5 A0A0A9XP81 B4MCL9 B4MIT0 B4JVQ1 A0A0J9RBW6 B4P4G9 Q6YKA8 Q08012 A0A3B0IZX0 B3NRK3 A0A224XWK1 A0A0P4VL81 A0A0V0GB86 A0A069DQL3 U5EXG7 A0A023F5F1 T1DNU8 A0A2M4CT90 A0A2M3Z8F8 A0A2M4AJ52 Q7PV64 A0A1I8PIL0 B3MGZ3 B4HQJ5 A0A1S3D1J4 A0A1W4VHZ6 D6WXI3 Q28ZT1 B4GGT9 T1E300 A0A1Q3EU64 A0A1L8DMA8 A0A1B6FSE9 A0A1B6LQ14 A0A0C9RMZ5 B4KMK9 K7IXW9 T1DIQ9 A0A067QZD3 A0A023EIK5 A0A232EQB5 A0A026WTT4 W8BBJ7 A0A0K8WEM4 A0A0A1X3H7 A0A034W8P6 A0A195FLT0 A0A158NEL3 A0A0J7LAC2 V9IHL5 A0A1J1J692 A0A0P6FKN8 A0A1W4WNL1 A0A087ZXM2 A0A195EN05 A0A0N0BKX1 V5GM45 A0A164Y7E8 A0A336KB09 A0A195BZU9 A0A151XIS8 A0A2A3EC90 A0A0N8DRV2 A0A2P2IA47 A0A2J7RGH4 A0A0P6IB62 T1IWZ8 E0VST4 A0A0M4MF07 J3JVP3 N6T8R7 A0A1Y1MVF9 A0A182L4P7 A0A195BUS7 T1HSZ9 A0A0P4WHY9 A0A0L7REL4 A0A087UW82 F4WRU7 A0A0P5XPK9 A0A131YIU8 A0A131XI35 L7M546 A0A1E1X7L1
A0A0L7LVH6 A0A212FIT9 A0A0K8TSK2 A0A2P8ZGQ7 A0A1B6E815 A0A1I8M7H0 A0A1S3D1F1 D3TPP1 A0A0L0C544 A0A146M7N5 A0A0A9XP81 B4MCL9 B4MIT0 B4JVQ1 A0A0J9RBW6 B4P4G9 Q6YKA8 Q08012 A0A3B0IZX0 B3NRK3 A0A224XWK1 A0A0P4VL81 A0A0V0GB86 A0A069DQL3 U5EXG7 A0A023F5F1 T1DNU8 A0A2M4CT90 A0A2M3Z8F8 A0A2M4AJ52 Q7PV64 A0A1I8PIL0 B3MGZ3 B4HQJ5 A0A1S3D1J4 A0A1W4VHZ6 D6WXI3 Q28ZT1 B4GGT9 T1E300 A0A1Q3EU64 A0A1L8DMA8 A0A1B6FSE9 A0A1B6LQ14 A0A0C9RMZ5 B4KMK9 K7IXW9 T1DIQ9 A0A067QZD3 A0A023EIK5 A0A232EQB5 A0A026WTT4 W8BBJ7 A0A0K8WEM4 A0A0A1X3H7 A0A034W8P6 A0A195FLT0 A0A158NEL3 A0A0J7LAC2 V9IHL5 A0A1J1J692 A0A0P6FKN8 A0A1W4WNL1 A0A087ZXM2 A0A195EN05 A0A0N0BKX1 V5GM45 A0A164Y7E8 A0A336KB09 A0A195BZU9 A0A151XIS8 A0A2A3EC90 A0A0N8DRV2 A0A2P2IA47 A0A2J7RGH4 A0A0P6IB62 T1IWZ8 E0VST4 A0A0M4MF07 J3JVP3 N6T8R7 A0A1Y1MVF9 A0A182L4P7 A0A195BUS7 T1HSZ9 A0A0P4WHY9 A0A0L7REL4 A0A087UW82 F4WRU7 A0A0P5XPK9 A0A131YIU8 A0A131XI35 L7M546 A0A1E1X7L1
Pubmed
26354079
23622113
26227816
22118469
26369729
29403074
+ More
25315136 20353571 26108605 26823975 25401762 17994087 22936249 17550304 12694293 8462097 8462098 10731132 12537572 12537569 9671493 12128212 18057021 27129103 26334808 25474469 12364791 14747013 17210077 18362917 19820115 15632085 24330624 20075255 24845553 24945155 28648823 24508170 30249741 24495485 25830018 25348373 21347285 20566863 22516182 23537049 28004739 20966253 21719571 26830274 28049606 25576852 28503490
25315136 20353571 26108605 26823975 25401762 17994087 22936249 17550304 12694293 8462097 8462098 10731132 12537572 12537569 9671493 12128212 18057021 27129103 26334808 25474469 12364791 14747013 17210077 18362917 19820115 15632085 24330624 20075255 24845553 24945155 28648823 24508170 30249741 24495485 25830018 25348373 21347285 20566863 22516182 23537049 28004739 20966253 21719571 26830274 28049606 25576852 28503490
EMBL
HQ540305
ADZ99023.1
KQ461155
KPJ08203.1
KQ459232
KPJ02732.1
+ More
GAIX01000554 JAA92006.1 ODYU01008453 SOQ52079.1 GDQN01003318 JAT87736.1 JTDY01000012 KOB79435.1 AGBW02008335 OWR53635.1 GDAI01000678 JAI16925.1 PYGN01000062 PSN55685.1 GEDC01011967 GEDC01003253 JAS25331.1 JAS34045.1 CCAG010016887 EZ423393 ADD19669.1 JRES01000890 KNC27518.1 GDHC01005523 GDHC01002856 JAQ13106.1 JAQ15773.1 GBHO01021097 GBHO01021067 GBHO01021036 GBHO01004926 JAG22507.1 JAG22537.1 JAG22568.1 JAG38678.1 CH940659 EDW71407.1 CH963719 EDW72019.1 CH916375 EDV98519.1 CM002911 KMY93517.1 KMY93518.1 CM000158 EDW90608.1 KRJ99318.1 KRJ99319.1 AY135137 AY135136 L12446 L13173 AY135053 AY135054 AY135055 AY135056 AY135057 AY135058 AY135059 AY135060 AY135061 AY135062 AY135063 AY135064 AY135065 AY135066 AY135067 AY135068 AY135069 AY135070 AY135071 AY135072 AY135073 AY135074 AE013599 AY061142 OUUW01000001 SPP73757.1 CH954179 EDV56155.1 KQS62728.1 GFTR01003556 JAW12870.1 GDKW01000721 JAI55874.1 GECL01000947 JAP05177.1 GBGD01002913 JAC85976.1 GANO01000963 JAB58908.1 GBBI01002343 JAC16369.1 GAMD01002555 JAA99035.1 GGFL01004213 MBW68391.1 GGFM01004068 MBW24819.1 GGFK01007480 MBW40801.1 AAAB01008986 EAA00404.4 CH902619 EDV37911.1 CH480816 EDW47730.1 KQ971361 EFA08848.1 CM000071 EAL25532.1 CH479183 EDW35709.1 GALA01000830 JAA94022.1 GFDL01016190 JAV18855.1 GFDF01006590 JAV07494.1 GECZ01016675 JAS53094.1 GEBQ01016660 GEBQ01014363 JAT23317.1 JAT25614.1 GBYB01008436 GBYB01008523 JAG78203.1 JAG78290.1 CH933808 EDW10856.1 GALA01000831 JAA94021.1 KK853076 KDR11728.1 GAPW01004335 JAC09263.1 NNAY01002790 OXU20555.1 KK107109 QOIP01000008 EZA59373.1 RLU19532.1 GAMC01012158 GAMC01012157 GAMC01012156 JAB94398.1 GDHF01027396 GDHF01026559 GDHF01025281 GDHF01018103 GDHF01003527 GDHF01002820 GDHF01000145 JAI24918.1 JAI25755.1 JAI27033.1 JAI34211.1 JAI48787.1 JAI49494.1 JAI52169.1 GBXI01011326 GBXI01010505 GBXI01008423 GBXI01002689 JAD02966.1 JAD03787.1 JAD05869.1 JAD11603.1 GAKP01008809 GAKP01008808 GAKP01008807 JAC50143.1 KQ981490 KYN41207.1 ADTU01013490 LBMM01000137 KMR04788.1 JR046105 AEY60137.1 CVRI01000074 CRL07912.1 GDIQ01055981 JAN38756.1 KQ978625 KYN29583.1 KQ435689 KOX81204.1 GALX01003292 JAB65174.1 LRGB01000930 KZS14957.1 UFQS01000117 UFQT01000117 SSW99973.1 SSX20353.1 KQ978501 KYM93451.1 KQ982080 KYQ60312.1 KZ288288 PBC29310.1 GDIP01007031 JAM96684.1 IACF01005313 LAB70899.1 NEVH01003784 PNF39920.1 GDIQ01023523 JAN71214.1 JH431636 DS235756 EEB16440.1 KR075866 ALE20585.1 BT127311 KB632061 AEE62273.1 ERL88402.1 APGK01039590 APGK01039591 APGK01039592 KB740972 ENN76619.1 GEZM01024109 GEZM01024108 JAV87966.1 KQ976401 KYM92374.1 ACPB03014463 GDRN01038395 GDRN01038393 JAI67255.1 KQ414608 KOC69392.1 KK121952 KFM81621.1 GL888292 EGI63104.1 GDIP01069114 JAM34601.1 GEDV01010079 JAP78478.1 GEFH01002549 JAP66032.1 GACK01005894 JAA59140.1 GFAC01003945 JAT95243.1
GAIX01000554 JAA92006.1 ODYU01008453 SOQ52079.1 GDQN01003318 JAT87736.1 JTDY01000012 KOB79435.1 AGBW02008335 OWR53635.1 GDAI01000678 JAI16925.1 PYGN01000062 PSN55685.1 GEDC01011967 GEDC01003253 JAS25331.1 JAS34045.1 CCAG010016887 EZ423393 ADD19669.1 JRES01000890 KNC27518.1 GDHC01005523 GDHC01002856 JAQ13106.1 JAQ15773.1 GBHO01021097 GBHO01021067 GBHO01021036 GBHO01004926 JAG22507.1 JAG22537.1 JAG22568.1 JAG38678.1 CH940659 EDW71407.1 CH963719 EDW72019.1 CH916375 EDV98519.1 CM002911 KMY93517.1 KMY93518.1 CM000158 EDW90608.1 KRJ99318.1 KRJ99319.1 AY135137 AY135136 L12446 L13173 AY135053 AY135054 AY135055 AY135056 AY135057 AY135058 AY135059 AY135060 AY135061 AY135062 AY135063 AY135064 AY135065 AY135066 AY135067 AY135068 AY135069 AY135070 AY135071 AY135072 AY135073 AY135074 AE013599 AY061142 OUUW01000001 SPP73757.1 CH954179 EDV56155.1 KQS62728.1 GFTR01003556 JAW12870.1 GDKW01000721 JAI55874.1 GECL01000947 JAP05177.1 GBGD01002913 JAC85976.1 GANO01000963 JAB58908.1 GBBI01002343 JAC16369.1 GAMD01002555 JAA99035.1 GGFL01004213 MBW68391.1 GGFM01004068 MBW24819.1 GGFK01007480 MBW40801.1 AAAB01008986 EAA00404.4 CH902619 EDV37911.1 CH480816 EDW47730.1 KQ971361 EFA08848.1 CM000071 EAL25532.1 CH479183 EDW35709.1 GALA01000830 JAA94022.1 GFDL01016190 JAV18855.1 GFDF01006590 JAV07494.1 GECZ01016675 JAS53094.1 GEBQ01016660 GEBQ01014363 JAT23317.1 JAT25614.1 GBYB01008436 GBYB01008523 JAG78203.1 JAG78290.1 CH933808 EDW10856.1 GALA01000831 JAA94021.1 KK853076 KDR11728.1 GAPW01004335 JAC09263.1 NNAY01002790 OXU20555.1 KK107109 QOIP01000008 EZA59373.1 RLU19532.1 GAMC01012158 GAMC01012157 GAMC01012156 JAB94398.1 GDHF01027396 GDHF01026559 GDHF01025281 GDHF01018103 GDHF01003527 GDHF01002820 GDHF01000145 JAI24918.1 JAI25755.1 JAI27033.1 JAI34211.1 JAI48787.1 JAI49494.1 JAI52169.1 GBXI01011326 GBXI01010505 GBXI01008423 GBXI01002689 JAD02966.1 JAD03787.1 JAD05869.1 JAD11603.1 GAKP01008809 GAKP01008808 GAKP01008807 JAC50143.1 KQ981490 KYN41207.1 ADTU01013490 LBMM01000137 KMR04788.1 JR046105 AEY60137.1 CVRI01000074 CRL07912.1 GDIQ01055981 JAN38756.1 KQ978625 KYN29583.1 KQ435689 KOX81204.1 GALX01003292 JAB65174.1 LRGB01000930 KZS14957.1 UFQS01000117 UFQT01000117 SSW99973.1 SSX20353.1 KQ978501 KYM93451.1 KQ982080 KYQ60312.1 KZ288288 PBC29310.1 GDIP01007031 JAM96684.1 IACF01005313 LAB70899.1 NEVH01003784 PNF39920.1 GDIQ01023523 JAN71214.1 JH431636 DS235756 EEB16440.1 KR075866 ALE20585.1 BT127311 KB632061 AEE62273.1 ERL88402.1 APGK01039590 APGK01039591 APGK01039592 KB740972 ENN76619.1 GEZM01024109 GEZM01024108 JAV87966.1 KQ976401 KYM92374.1 ACPB03014463 GDRN01038395 GDRN01038393 JAI67255.1 KQ414608 KOC69392.1 KK121952 KFM81621.1 GL888292 EGI63104.1 GDIP01069114 JAM34601.1 GEDV01010079 JAP78478.1 GEFH01002549 JAP66032.1 GACK01005894 JAA59140.1 GFAC01003945 JAT95243.1
Proteomes
UP000053240
UP000053268
UP000037510
UP000007151
UP000245037
UP000095301
+ More
UP000079169 UP000092444 UP000037069 UP000008792 UP000007798 UP000001070 UP000002282 UP000000803 UP000268350 UP000008711 UP000007062 UP000095300 UP000007801 UP000001292 UP000192221 UP000007266 UP000001819 UP000008744 UP000009192 UP000002358 UP000027135 UP000215335 UP000053097 UP000279307 UP000078541 UP000005205 UP000036403 UP000183832 UP000192223 UP000005203 UP000078492 UP000053105 UP000076858 UP000078542 UP000075809 UP000242457 UP000235965 UP000009046 UP000030742 UP000019118 UP000075882 UP000078540 UP000015103 UP000053825 UP000054359 UP000007755
UP000079169 UP000092444 UP000037069 UP000008792 UP000007798 UP000001070 UP000002282 UP000000803 UP000268350 UP000008711 UP000007062 UP000095300 UP000007801 UP000001292 UP000192221 UP000007266 UP000001819 UP000008744 UP000009192 UP000002358 UP000027135 UP000215335 UP000053097 UP000279307 UP000078541 UP000005205 UP000036403 UP000183832 UP000192223 UP000005203 UP000078492 UP000053105 UP000076858 UP000078542 UP000075809 UP000242457 UP000235965 UP000009046 UP000030742 UP000019118 UP000075882 UP000078540 UP000015103 UP000053825 UP000054359 UP000007755
Interpro
Gene 3D
ProteinModelPortal
F2XHI6
A0A194QRR0
A0A194QB94
S4PH84
A0A2H1WG69
A0A1E1WLK8
+ More
A0A0L7LVH6 A0A212FIT9 A0A0K8TSK2 A0A2P8ZGQ7 A0A1B6E815 A0A1I8M7H0 A0A1S3D1F1 D3TPP1 A0A0L0C544 A0A146M7N5 A0A0A9XP81 B4MCL9 B4MIT0 B4JVQ1 A0A0J9RBW6 B4P4G9 Q6YKA8 Q08012 A0A3B0IZX0 B3NRK3 A0A224XWK1 A0A0P4VL81 A0A0V0GB86 A0A069DQL3 U5EXG7 A0A023F5F1 T1DNU8 A0A2M4CT90 A0A2M3Z8F8 A0A2M4AJ52 Q7PV64 A0A1I8PIL0 B3MGZ3 B4HQJ5 A0A1S3D1J4 A0A1W4VHZ6 D6WXI3 Q28ZT1 B4GGT9 T1E300 A0A1Q3EU64 A0A1L8DMA8 A0A1B6FSE9 A0A1B6LQ14 A0A0C9RMZ5 B4KMK9 K7IXW9 T1DIQ9 A0A067QZD3 A0A023EIK5 A0A232EQB5 A0A026WTT4 W8BBJ7 A0A0K8WEM4 A0A0A1X3H7 A0A034W8P6 A0A195FLT0 A0A158NEL3 A0A0J7LAC2 V9IHL5 A0A1J1J692 A0A0P6FKN8 A0A1W4WNL1 A0A087ZXM2 A0A195EN05 A0A0N0BKX1 V5GM45 A0A164Y7E8 A0A336KB09 A0A195BZU9 A0A151XIS8 A0A2A3EC90 A0A0N8DRV2 A0A2P2IA47 A0A2J7RGH4 A0A0P6IB62 T1IWZ8 E0VST4 A0A0M4MF07 J3JVP3 N6T8R7 A0A1Y1MVF9 A0A182L4P7 A0A195BUS7 T1HSZ9 A0A0P4WHY9 A0A0L7REL4 A0A087UW82 F4WRU7 A0A0P5XPK9 A0A131YIU8 A0A131XI35 L7M546 A0A1E1X7L1
A0A0L7LVH6 A0A212FIT9 A0A0K8TSK2 A0A2P8ZGQ7 A0A1B6E815 A0A1I8M7H0 A0A1S3D1F1 D3TPP1 A0A0L0C544 A0A146M7N5 A0A0A9XP81 B4MCL9 B4MIT0 B4JVQ1 A0A0J9RBW6 B4P4G9 Q6YKA8 Q08012 A0A3B0IZX0 B3NRK3 A0A224XWK1 A0A0P4VL81 A0A0V0GB86 A0A069DQL3 U5EXG7 A0A023F5F1 T1DNU8 A0A2M4CT90 A0A2M3Z8F8 A0A2M4AJ52 Q7PV64 A0A1I8PIL0 B3MGZ3 B4HQJ5 A0A1S3D1J4 A0A1W4VHZ6 D6WXI3 Q28ZT1 B4GGT9 T1E300 A0A1Q3EU64 A0A1L8DMA8 A0A1B6FSE9 A0A1B6LQ14 A0A0C9RMZ5 B4KMK9 K7IXW9 T1DIQ9 A0A067QZD3 A0A023EIK5 A0A232EQB5 A0A026WTT4 W8BBJ7 A0A0K8WEM4 A0A0A1X3H7 A0A034W8P6 A0A195FLT0 A0A158NEL3 A0A0J7LAC2 V9IHL5 A0A1J1J692 A0A0P6FKN8 A0A1W4WNL1 A0A087ZXM2 A0A195EN05 A0A0N0BKX1 V5GM45 A0A164Y7E8 A0A336KB09 A0A195BZU9 A0A151XIS8 A0A2A3EC90 A0A0N8DRV2 A0A2P2IA47 A0A2J7RGH4 A0A0P6IB62 T1IWZ8 E0VST4 A0A0M4MF07 J3JVP3 N6T8R7 A0A1Y1MVF9 A0A182L4P7 A0A195BUS7 T1HSZ9 A0A0P4WHY9 A0A0L7REL4 A0A087UW82 F4WRU7 A0A0P5XPK9 A0A131YIU8 A0A131XI35 L7M546 A0A1E1X7L1
PDB
1GRI
E-value=7.50089e-68,
Score=649
Ontologies
PATHWAY
GO
GO:0007265
GO:0016301
GO:0007614
GO:0070374
GO:0005118
GO:0045500
GO:0032991
GO:0001784
GO:1904263
GO:0008543
GO:0048260
GO:0045793
GO:0007173
GO:0007426
GO:0005154
GO:0008293
GO:0007476
GO:0005070
GO:0030707
GO:0048010
GO:0007427
GO:0005886
GO:0008355
GO:0007465
GO:0008306
GO:0005654
GO:0005737
GO:0005515
GO:0006511
GO:0016579
GO:0006397
GO:0005524
GO:0004812
GO:0006418
GO:0003723
GO:0046872
GO:0003824
PANTHER
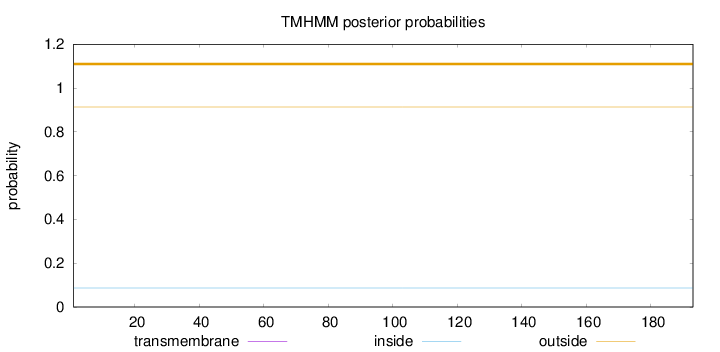
Topology
Subcellular location
Membrane
Length:
193
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00058
Exp number, first 60 AAs:
0
Total prob of N-in:
0.08698
outside
1 - 193
Population Genetic Test Statistics
Pi
22.846764
Theta
203.64551
Tajima's D
-0.895296
CLR
0.575198
CSRT
0.155942202889855
Interpretation
Uncertain
