Pre Gene Modal
BGIBMGA007318
Annotation
PREDICTED:_uncharacterized_protein_LOC101735385_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.061
Sequence
CDS
ATGGCTACCGCGAAAGACACGTCTACTTTCTTTCTGGAGGCAAATGCAAAAGAATTCGATAGTGTATTAAAGTTGTATCCTCAGGCGATTAAACTAAAAGCTGAACGTAAAACAAAAAGACCAGATGAGCTCATAAAATTGGACAATTGGTATCAAAATGAACTTCCCAAGAAGATCAAATCGCGGGGTAAAGATGCGCACATGATCCACGAAGAACTTGTCCAGCTCATGAAATGGAAACAGGCCAGAGGAAAATTCTACCCGCAGTTGTCGTATCTGATAAAAGTGAACACGCCACGAGCTGTGATGCAAGAGACGAAAAAGGCCTTCCGCAAACTGCCCAATATCGAATCCGCGATGACCGCTCTAAGCAATCTCAAAGGCGTGGGAACGGCCACAGCATCAGCGTTATTAGCTGCCGCTAGTCCAGAAATAGCACCGTTTATGGCTGACGAATGCGTCCAGGCGATCCCCGAAATGGAAGGCAGCGATTACACAGCAAGGGAATACCTCAATTTCGAACAAAACGGCTGCGGCAAGAAATGGTTTCCACACATGGTAGAGCTGGCCCTCTGGACGCACAACATTGTATCGGACTTACAACCACAGTTATTAGGGAAAGAGCCCAACAACCGTCCGGCTGCAAACGGCGGCTCCCCCCTTCCCAGTGACGAGAGCAACTTAGAGCCCCCAGCGACAAACGGTAAACTCGTAACGGACTGCGTGAATGAGGATACCACGACGTCGTGCACAGAAGACTCGATGGACGCGAAAGCCGCATCACCCTCCACCCCCGCTACCCCTGCCACTCCAGCGTCACCCTCAGACAACTCAGACTCTGCAGTCAGCACGCCACGAACTAAAAGACAATTGGAGGAAGGTAGTTCGGCGGAAGAAAACTCACTGGGAGACTCGTCTGAGGCCACGCTACCTAACATCGCAAAGAAACTAAGGAAAGCCACACACTGA
Protein
MATAKDTSTFFLEANAKEFDSVLKLYPQAIKLKAERKTKRPDELIKLDNWYQNELPKKIKSRGKDAHMIHEELVQLMKWKQARGKFYPQLSYLIKVNTPRAVMQETKKAFRKLPNIESAMTALSNLKGVGTATASALLAAASPEIAPFMADECVQAIPEMEGSDYTAREYLNFEQNGCGKKWFPHMVELALWTHNIVSDLQPQLLGKEPNNRPAANGGSPLPSDESNLEPPATNGKLVTDCVNEDTTTSCTEDSMDAKAASPSTPATPATPASPSDNSDSAVSTPRTKRQLEEGSSAEENSLGDSSEATLPNIAKKLRKATH
Summary
Uniprot
H9JCS1
A0A2A4K743
A0A3S2LE56
A0A194QB08
A0A2H1WIL2
A0A1E1WHZ4
+ More
S4P851 A0A194QXA4 A0A2A4K8A4 A0A154PIZ8 A0A151XIS1 A0A026WW71 A0A0L7RF95 A0A1E1WRU6 E9IJA9 A0A195FKL7 V9IJ05 A0A087ZXD3 E2BG85 A0A0J7LA47 A0A195BXY0 A0A195BWM5 A0A158NEL4 E2AFN1 F4WRU5 A0A0T6B735 A0A1W4WR95 A0A1L8D882 A0A1Y1LEX2 A0A1B6JEI8 A0A1B6G203 D6WX85 A0A1B6E3K4 V5H5C3 A0A1L8D8F3 A0A232EXG4 A0A1B6MK68 E0VST2 A0A0P4VMN9 A0A0V0GAE6 A0A023F4E8 A0A224XGM0 A0A069DYJ1 A0A0A9W8I5 A0A2R7WSY1 A0A067QWF8 T1HB91 Q177C3 B0W349 A0A0M9ABQ6 A0A1A9YMH1 A0A1B0BDK6 A0A2S2P8W3 J9JPY5 T1PA83 A0A0M4EV10 A0A1B0FB05 A0A1B0A829 A0A182GLD6 A0A182HB85 A0A1S4FCN5 B4L678 A0A336L755 A0A034WVE2 A0A0K8W0J5 A9YGK3 A9YGJ9 A0A0A1WHW9 F6J9S0 F6J9Q0 A9YGJ8 B4JM15 A0A1J1J6X4 B3MSC1 Q9VYV4 B4Q1S0 B3NUA0 B4R364 B4IK52 A0A1B6GM20 A0A0L7LWA6 B4MT18 K7IXX0
S4P851 A0A194QXA4 A0A2A4K8A4 A0A154PIZ8 A0A151XIS1 A0A026WW71 A0A0L7RF95 A0A1E1WRU6 E9IJA9 A0A195FKL7 V9IJ05 A0A087ZXD3 E2BG85 A0A0J7LA47 A0A195BXY0 A0A195BWM5 A0A158NEL4 E2AFN1 F4WRU5 A0A0T6B735 A0A1W4WR95 A0A1L8D882 A0A1Y1LEX2 A0A1B6JEI8 A0A1B6G203 D6WX85 A0A1B6E3K4 V5H5C3 A0A1L8D8F3 A0A232EXG4 A0A1B6MK68 E0VST2 A0A0P4VMN9 A0A0V0GAE6 A0A023F4E8 A0A224XGM0 A0A069DYJ1 A0A0A9W8I5 A0A2R7WSY1 A0A067QWF8 T1HB91 Q177C3 B0W349 A0A0M9ABQ6 A0A1A9YMH1 A0A1B0BDK6 A0A2S2P8W3 J9JPY5 T1PA83 A0A0M4EV10 A0A1B0FB05 A0A1B0A829 A0A182GLD6 A0A182HB85 A0A1S4FCN5 B4L678 A0A336L755 A0A034WVE2 A0A0K8W0J5 A9YGK3 A9YGJ9 A0A0A1WHW9 F6J9S0 F6J9Q0 A9YGJ8 B4JM15 A0A1J1J6X4 B3MSC1 Q9VYV4 B4Q1S0 B3NUA0 B4R364 B4IK52 A0A1B6GM20 A0A0L7LWA6 B4MT18 K7IXX0
Pubmed
19121390
26354079
23622113
24508170
30249741
21282665
+ More
20798317 21347285 21719571 28004739 18362917 19820115 28648823 20566863 27129103 25474469 26334808 25401762 26823975 24845553 17510324 25315136 26483478 17994087 18057021 25348373 17989248 21383965 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26227816 20075255
20798317 21347285 21719571 28004739 18362917 19820115 28648823 20566863 27129103 25474469 26334808 25401762 26823975 24845553 17510324 25315136 26483478 17994087 18057021 25348373 17989248 21383965 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26227816 20075255
EMBL
BABH01041510
BABH01041511
BABH01041512
NWSH01000075
PCG79899.1
RSAL01000029
+ More
RVE51784.1 KQ459232 KPJ02728.1 ODYU01008918 SOQ52913.1 GDQN01004452 JAT86602.1 GAIX01004304 JAA88256.1 KQ461155 KPJ08206.1 PCG79900.1 KQ434931 KZC11815.1 KQ982080 KYQ60309.1 KK107109 QOIP01000008 EZA59374.1 RLU19531.1 KQ414608 KOC69391.1 GDQN01011363 GDQN01001291 JAT79691.1 JAT89763.1 GL763764 EFZ19329.1 KQ981490 KYN41210.1 JR049690 AEY61040.1 GL448116 EFN85293.1 LBMM01000137 KMR04787.1 KQ978501 KYM93452.1 KQ976401 KYM92373.1 ADTU01013495 GL439118 EFN67782.1 GL888292 EGI63102.1 LJIG01009475 KRT83006.1 GFDF01011436 JAV02648.1 GEZM01059539 GEZM01059538 JAV71438.1 GECU01010157 JAS97549.1 GECZ01013292 JAS56477.1 KQ971361 EFA08023.2 GEDC01013026 GEDC01004803 JAS24272.1 JAS32495.1 GALX01000468 JAB67998.1 GFDF01011447 JAV02637.1 NNAY01001763 OXU22987.1 GEBQ01003666 JAT36311.1 DS235756 EEB16438.1 GDKW01003002 JAI53593.1 GECL01001221 JAP04903.1 GBBI01002670 JAC16042.1 GFTR01004790 JAW11636.1 GBGD01002300 JAC86589.1 GBHO01040786 GBRD01006847 GBRD01006846 GDHC01003642 JAG02818.1 JAG58974.1 JAQ14987.1 KK855416 PTY22461.1 KK853286 KDR08958.1 ACPB03014463 CH477378 EAT42269.1 DS231830 EDS30757.1 KQ435689 KOX81210.1 JXJN01012514 GGMR01013284 MBY25903.1 ABLF02033111 KA645050 AFP59679.1 CP012528 ALC49022.1 CCAG010001425 JXUM01071762 JXUM01071763 JXUM01071764 JXUM01071765 JXUM01071766 JXUM01071767 JXUM01032539 JXUM01032540 CH933812 EDW05874.1 KRG07066.1 KRG07067.1 UFQS01001834 UFQT01001834 SSX12493.1 SSX31936.1 GAKP01001229 JAC57723.1 GDHF01007974 JAI44340.1 EU216882 EU216883 EU216886 EU216888 GQ306816 GQ306818 GQ306820 GQ306821 GQ306822 ABW91839.1 ABW91840.1 ABW91843.1 ABW91845.1 ADG50261.1 ADG50263.1 ADG50265.1 ADG50266.1 ADG50267.1 EU216878 EU216879 EU216880 EU216881 EU216884 EU216885 EU216887 EU216889 GQ306817 GQ306819 GQ306823 GQ306824 GQ306825 GQ306826 GQ306827 ABW91835.1 ABW91836.1 ABW91837.1 ABW91838.1 ABW91841.1 ABW91842.1 ABW91844.1 ABW91846.1 ADG50262.1 ADG50264.1 ADG50268.1 ADG50269.1 ADG50270.1 ADG50271.1 ADG50272.1 GBXI01015653 JAC98638.1 GQ311890 GQ311892 GQ311893 GQ311894 GQ311895 GQ311896 GQ311897 GQ311898 GQ311900 GQ311901 GQ311902 GQ311903 GQ311904 GQ311905 GQ311906 GQ311907 GQ311908 GQ311910 GQ311911 ADG50239.1 ADG50241.1 ADG50242.1 ADG50243.1 ADG50244.1 ADG50245.1 ADG50246.1 ADG50247.1 ADG50249.1 ADG50250.1 ADG50251.1 ADG50252.1 ADG50253.1 ADG50254.1 ADG50255.1 ADG50256.1 ADG50257.1 ADG50259.1 ADG50260.1 GQ311891 GQ311899 GQ311909 ADG50240.1 ADG50248.1 ADG50258.1 EU216877 ABW91834.1 CH916371 EDV91776.1 CVRI01000074 CRL08135.1 CH902622 EDV34676.1 KPU75240.1 AE014298 BT001379 AAF48080.1 AAF48081.1 AAG22344.1 AAN09295.1 AAN71134.1 ADV37661.1 ADV37662.1 ADV37663.1 AGB95302.1 AHN59606.1 CM000162 EDX02495.1 CH954180 EDV46015.1 KQS29789.1 KQS29790.1 KQS29791.1 KQS29792.1 CM000366 EDX17666.1 CH480851 EDW51424.1 GECZ01006267 JAS63502.1 JTDY01000012 KOB79441.1 CH963851 EDW75257.1
RVE51784.1 KQ459232 KPJ02728.1 ODYU01008918 SOQ52913.1 GDQN01004452 JAT86602.1 GAIX01004304 JAA88256.1 KQ461155 KPJ08206.1 PCG79900.1 KQ434931 KZC11815.1 KQ982080 KYQ60309.1 KK107109 QOIP01000008 EZA59374.1 RLU19531.1 KQ414608 KOC69391.1 GDQN01011363 GDQN01001291 JAT79691.1 JAT89763.1 GL763764 EFZ19329.1 KQ981490 KYN41210.1 JR049690 AEY61040.1 GL448116 EFN85293.1 LBMM01000137 KMR04787.1 KQ978501 KYM93452.1 KQ976401 KYM92373.1 ADTU01013495 GL439118 EFN67782.1 GL888292 EGI63102.1 LJIG01009475 KRT83006.1 GFDF01011436 JAV02648.1 GEZM01059539 GEZM01059538 JAV71438.1 GECU01010157 JAS97549.1 GECZ01013292 JAS56477.1 KQ971361 EFA08023.2 GEDC01013026 GEDC01004803 JAS24272.1 JAS32495.1 GALX01000468 JAB67998.1 GFDF01011447 JAV02637.1 NNAY01001763 OXU22987.1 GEBQ01003666 JAT36311.1 DS235756 EEB16438.1 GDKW01003002 JAI53593.1 GECL01001221 JAP04903.1 GBBI01002670 JAC16042.1 GFTR01004790 JAW11636.1 GBGD01002300 JAC86589.1 GBHO01040786 GBRD01006847 GBRD01006846 GDHC01003642 JAG02818.1 JAG58974.1 JAQ14987.1 KK855416 PTY22461.1 KK853286 KDR08958.1 ACPB03014463 CH477378 EAT42269.1 DS231830 EDS30757.1 KQ435689 KOX81210.1 JXJN01012514 GGMR01013284 MBY25903.1 ABLF02033111 KA645050 AFP59679.1 CP012528 ALC49022.1 CCAG010001425 JXUM01071762 JXUM01071763 JXUM01071764 JXUM01071765 JXUM01071766 JXUM01071767 JXUM01032539 JXUM01032540 CH933812 EDW05874.1 KRG07066.1 KRG07067.1 UFQS01001834 UFQT01001834 SSX12493.1 SSX31936.1 GAKP01001229 JAC57723.1 GDHF01007974 JAI44340.1 EU216882 EU216883 EU216886 EU216888 GQ306816 GQ306818 GQ306820 GQ306821 GQ306822 ABW91839.1 ABW91840.1 ABW91843.1 ABW91845.1 ADG50261.1 ADG50263.1 ADG50265.1 ADG50266.1 ADG50267.1 EU216878 EU216879 EU216880 EU216881 EU216884 EU216885 EU216887 EU216889 GQ306817 GQ306819 GQ306823 GQ306824 GQ306825 GQ306826 GQ306827 ABW91835.1 ABW91836.1 ABW91837.1 ABW91838.1 ABW91841.1 ABW91842.1 ABW91844.1 ABW91846.1 ADG50262.1 ADG50264.1 ADG50268.1 ADG50269.1 ADG50270.1 ADG50271.1 ADG50272.1 GBXI01015653 JAC98638.1 GQ311890 GQ311892 GQ311893 GQ311894 GQ311895 GQ311896 GQ311897 GQ311898 GQ311900 GQ311901 GQ311902 GQ311903 GQ311904 GQ311905 GQ311906 GQ311907 GQ311908 GQ311910 GQ311911 ADG50239.1 ADG50241.1 ADG50242.1 ADG50243.1 ADG50244.1 ADG50245.1 ADG50246.1 ADG50247.1 ADG50249.1 ADG50250.1 ADG50251.1 ADG50252.1 ADG50253.1 ADG50254.1 ADG50255.1 ADG50256.1 ADG50257.1 ADG50259.1 ADG50260.1 GQ311891 GQ311899 GQ311909 ADG50240.1 ADG50248.1 ADG50258.1 EU216877 ABW91834.1 CH916371 EDV91776.1 CVRI01000074 CRL08135.1 CH902622 EDV34676.1 KPU75240.1 AE014298 BT001379 AAF48080.1 AAF48081.1 AAG22344.1 AAN09295.1 AAN71134.1 ADV37661.1 ADV37662.1 ADV37663.1 AGB95302.1 AHN59606.1 CM000162 EDX02495.1 CH954180 EDV46015.1 KQS29789.1 KQS29790.1 KQS29791.1 KQS29792.1 CM000366 EDX17666.1 CH480851 EDW51424.1 GECZ01006267 JAS63502.1 JTDY01000012 KOB79441.1 CH963851 EDW75257.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000053240
UP000076502
+ More
UP000075809 UP000053097 UP000279307 UP000053825 UP000078541 UP000005203 UP000008237 UP000036403 UP000078542 UP000078540 UP000005205 UP000000311 UP000007755 UP000192223 UP000007266 UP000215335 UP000009046 UP000027135 UP000015103 UP000008820 UP000002320 UP000053105 UP000092443 UP000092460 UP000007819 UP000095301 UP000092553 UP000092444 UP000092445 UP000069940 UP000009192 UP000001070 UP000183832 UP000007801 UP000000803 UP000002282 UP000008711 UP000000304 UP000001292 UP000037510 UP000007798 UP000002358
UP000075809 UP000053097 UP000279307 UP000053825 UP000078541 UP000005203 UP000008237 UP000036403 UP000078542 UP000078540 UP000005205 UP000000311 UP000007755 UP000192223 UP000007266 UP000215335 UP000009046 UP000027135 UP000015103 UP000008820 UP000002320 UP000053105 UP000092443 UP000092460 UP000007819 UP000095301 UP000092553 UP000092444 UP000092445 UP000069940 UP000009192 UP000001070 UP000183832 UP000007801 UP000000803 UP000002282 UP000008711 UP000000304 UP000001292 UP000037510 UP000007798 UP000002358
Interpro
IPR016024
ARM-type_fold
SUPFAM
SSF48371
SSF48371
ProteinModelPortal
H9JCS1
A0A2A4K743
A0A3S2LE56
A0A194QB08
A0A2H1WIL2
A0A1E1WHZ4
+ More
S4P851 A0A194QXA4 A0A2A4K8A4 A0A154PIZ8 A0A151XIS1 A0A026WW71 A0A0L7RF95 A0A1E1WRU6 E9IJA9 A0A195FKL7 V9IJ05 A0A087ZXD3 E2BG85 A0A0J7LA47 A0A195BXY0 A0A195BWM5 A0A158NEL4 E2AFN1 F4WRU5 A0A0T6B735 A0A1W4WR95 A0A1L8D882 A0A1Y1LEX2 A0A1B6JEI8 A0A1B6G203 D6WX85 A0A1B6E3K4 V5H5C3 A0A1L8D8F3 A0A232EXG4 A0A1B6MK68 E0VST2 A0A0P4VMN9 A0A0V0GAE6 A0A023F4E8 A0A224XGM0 A0A069DYJ1 A0A0A9W8I5 A0A2R7WSY1 A0A067QWF8 T1HB91 Q177C3 B0W349 A0A0M9ABQ6 A0A1A9YMH1 A0A1B0BDK6 A0A2S2P8W3 J9JPY5 T1PA83 A0A0M4EV10 A0A1B0FB05 A0A1B0A829 A0A182GLD6 A0A182HB85 A0A1S4FCN5 B4L678 A0A336L755 A0A034WVE2 A0A0K8W0J5 A9YGK3 A9YGJ9 A0A0A1WHW9 F6J9S0 F6J9Q0 A9YGJ8 B4JM15 A0A1J1J6X4 B3MSC1 Q9VYV4 B4Q1S0 B3NUA0 B4R364 B4IK52 A0A1B6GM20 A0A0L7LWA6 B4MT18 K7IXX0
S4P851 A0A194QXA4 A0A2A4K8A4 A0A154PIZ8 A0A151XIS1 A0A026WW71 A0A0L7RF95 A0A1E1WRU6 E9IJA9 A0A195FKL7 V9IJ05 A0A087ZXD3 E2BG85 A0A0J7LA47 A0A195BXY0 A0A195BWM5 A0A158NEL4 E2AFN1 F4WRU5 A0A0T6B735 A0A1W4WR95 A0A1L8D882 A0A1Y1LEX2 A0A1B6JEI8 A0A1B6G203 D6WX85 A0A1B6E3K4 V5H5C3 A0A1L8D8F3 A0A232EXG4 A0A1B6MK68 E0VST2 A0A0P4VMN9 A0A0V0GAE6 A0A023F4E8 A0A224XGM0 A0A069DYJ1 A0A0A9W8I5 A0A2R7WSY1 A0A067QWF8 T1HB91 Q177C3 B0W349 A0A0M9ABQ6 A0A1A9YMH1 A0A1B0BDK6 A0A2S2P8W3 J9JPY5 T1PA83 A0A0M4EV10 A0A1B0FB05 A0A1B0A829 A0A182GLD6 A0A182HB85 A0A1S4FCN5 B4L678 A0A336L755 A0A034WVE2 A0A0K8W0J5 A9YGK3 A9YGJ9 A0A0A1WHW9 F6J9S0 F6J9Q0 A9YGJ8 B4JM15 A0A1J1J6X4 B3MSC1 Q9VYV4 B4Q1S0 B3NUA0 B4R364 B4IK52 A0A1B6GM20 A0A0L7LWA6 B4MT18 K7IXX0
Ontologies
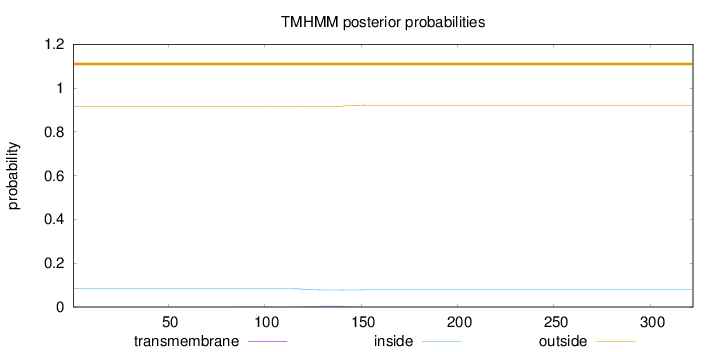
Topology
Length:
322
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.12735
Exp number, first 60 AAs:
0
Total prob of N-in:
0.08342
outside
1 - 322
Population Genetic Test Statistics
Pi
243.859738
Theta
197.77174
Tajima's D
0.6825
CLR
0.224577
CSRT
0.566471676416179
Interpretation
Uncertain
