Gene
KWMTBOMO10244
Annotation
PREDICTED:_protein_asunder-like_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.057 PlasmaMembrane Reliability : 1.364
Sequence
CDS
ATGAATTCCCTTTTCCCTGTAAATCATAAAACAATATTTGTTCTTGATCACACACCGTACTTTGGAATATCTTCAGAATACCCAATAGATTTTGATGTATCGAAAGGCCGAGGTACCGGTCAGGTTCCTTTACCTCCAATCTGTAAATCATTGTGGACTTGCAGTGTGGAAGCTGCGGTAGAGTACTGTCGTATAGTGTGGGATTTATTTCCTGAAAGCAAATTAGTGAAATTTGTAGTGAGTGATTCTGTAGCGCATATATTAAACACTTGGGCGGCATTACAACAAAATTTAACTCATGTCCTGAATGGGTTGTGTCTTGTTGGACCAGTACGTCGTGGTGCTGGAGGGGATGTTGTCGGACTCTGTGCAGCCATCGAAGCACTGGGTGAAGCATCACCAGAGCAGATATCACGCACATCAACCGATAGACATTTTCAAAATAGAGGACGGATAATATGTATTACAAGTGCAAGAGATGATGATTCAATAAGGAGCTTAGCAGAAATAGCTTTAAACACTTTGATTCAACAAAACAAAAAAGCTTCCATAGTCCAACCAAACAGCAGTAGAGATTCTCCTAATGCAACACCTCAATTATTAGTAACACATTACTGCCATCTAATCATCATAAATGTCACACCAGAGAAAGCTGAAACCAGAGTTAACAATCAACAGCTCACAGAGATGGGCGGTGGTCTGATGGGCGTCGAAGTAATAGTATCACGAGCTAGCGCATTAGCTAGCTTGTTGGGTCGTCTGGTACTACCGCATTACAAATTATCAACTACCACAGTTACTGGAATTCCCATGAAGGAAGAACAAAACGCTAGTTCCAGTGCTAATTACGATGTAGAAATTGTCCATTCCATCGATGCTCATGCTGGCTTGCGAGCTTCACAAACTGCTGCCGAAGCCGCTGAGTCTGTGGTGCTACGTTGGTGGACGCCGCGCGGCGCGGAGGGGGGCGCGGGCGGCTGCACGGGTCCGCTGTGGCGCGTCACGCCCGCCGACGTCGCCAGCCGCCCCTCCGCCTGTCTCGTCAACTTCCTGCTCAACGGGCGGTCGGTCACGCTCGAAGTGCCCAGGAAAGGTGGGGGTAGATCAGCGTCGCACGTGCTGGCGGCCCAGGGGGGCGAGCTGTGGATCGGCGCCCTGGGGGGTCGGACCACGTACGACGATCCCCCCGCGCTCTCCGAGGGAGCCGGTGGACGCGTCACAGACTACAGAATAAAAGAGTTTGGCGCACTGATGCAGCAAAATAAGCTGCACCCACTCCGCGGGGCGGGGTCCAACGCACGCGCGAGGTCTCGGCTGCAAAGGCACACGACATATTGGCCCCTGACGATATCCTCAACGCTCATATTCAATTTAGGATCAGTGATGGAACCGTTAACGAGGCTGGTCGTGCAAGAATCATTAACTGACGAAGAGGTGGATCAGTGTCGTAGCGTCCTGCTGTCGTTATTAAGTATGGAATCTCGCCATGAATTCCCGGCCGCGCAGCTGCCTTCGAATCTAAGGGGCAAATCGAGCGGTCGTCGCGAAGAACAATGGCGTCAAGTTTGGGGTGAATTGGAAGCGTTATTACGAGCGTATTCGAACTCCTCCGCCCATAGAACGCTACTGAGAGTCCTGCTAGAATGTCGGGCACACAATAACCACGGCGCAGACGGTGAATCTCGAGCAACGGTGATCCGAGCGACCACCGACTCCCCACTGTCGCCGGTGGGCGCAGGTAATGGCAGCGCCAGTAGTGCAAGTGGAGACGTCTGGGCTCCCAGGAACGTTCTGGACGCGTTACTGGGGGCGCCGCGTCCCGCGCGCAGACTCGACTTCGCCGGCAGGATGGCGGCCGGAAACAGGGTGGCCATCTTGTACCCGCAGCTGCAGCAGGACGTGGAAACCCAACACTGA
Protein
MNSLFPVNHKTIFVLDHTPYFGISSEYPIDFDVSKGRGTGQVPLPPICKSLWTCSVEAAVEYCRIVWDLFPESKLVKFVVSDSVAHILNTWAALQQNLTHVLNGLCLVGPVRRGAGGDVVGLCAAIEALGEASPEQISRTSTDRHFQNRGRIICITSARDDDSIRSLAEIALNTLIQQNKKASIVQPNSSRDSPNATPQLLVTHYCHLIIINVTPEKAETRVNNQQLTEMGGGLMGVEVIVSRASALASLLGRLVLPHYKLSTTTVTGIPMKEEQNASSSANYDVEIVHSIDAHAGLRASQTAAEAAESVVLRWWTPRGAEGGAGGCTGPLWRVTPADVASRPSACLVNFLLNGRSVTLEVPRKGGGRSASHVLAAQGGELWIGALGGRTTYDDPPALSEGAGGRVTDYRIKEFGALMQQNKLHPLRGAGSNARARSRLQRHTTYWPLTISSTLIFNLGSVMEPLTRLVVQESLTDEEVDQCRSVLLSLLSMESRHEFPAAQLPSNLRGKSSGRREEQWRQVWGELEALLRAYSNSSAHRTLLRVLLECRAHNNHGADGESRATVIRATTDSPLSPVGAGNGSASSASGDVWAPRNVLDALLGAPRPARRLDFAGRMAAGNRVAILYPQLQQDVETQH
Summary
Uniprot
A0A194QB89
A0A194QRU2
A0A2A4K6L8
A0A1W4X207
A0A1Y1K9K7
D6WX84
+ More
A0A2J7RGI8 A0A2J7RGG8 A0A1B6F841 A0A0C9RLQ9 E2AFM9 E2BG87 A0A158NEL6 A0A310SCX5 A0A154PK27 A0A2A3EC09 A0A0L7REY6 A0A1W4WR92 A0A0N0BKX5 A0A087ZXM1 A0A026WUN4 E0VUX7 E9IJB0 A0A0C9QD72 A0A195FLJ9 K7IYF5 A0A151XJ50 A0A232F2J8 A0A195BUW4 F4WRU4 A0A195BZW8 A0A195EN35 A0A1B6JEH0 A0A1B0D8A5 A0A2P8Y5Z3 A0A1B0CLA7 A0A336MX73 A0A336MS44 Q16N82 A0A3L8DGG3 A0A1Q3F0S6 A0A0J7KJK2 B0WR47 A0A182H4U1 A0A182R7L4 A0A084W9G5 A0A182MRE3 A0A182PEI2 A0A182W2L4 A0A182JID5 A0A182NG84 A0A182XLY0 Q7PTK3 A0A182HS30 A0A182XVD6 N6UCT2 A0A182US67 A0A182TSC2 A0A146LYB8 A0A0A9WSZ0 A0A182TX77 A0A0K8SVE8 T1J846 A0A224YNK6 A0A182FPZ3 D3TPL8 A0A1A9Z9U0 A0A2S2R0W1 A0A1B0FCI7 A0A1A9VG93 A0A131YFF8 A0A1E1XBT0 A0A131XC71 W5JUQ4 A0A2R5LJL0 A0A293MD54 J9K7L3 A0A2H8TFN9 A0A1B0BW91 A0A087T509 A0A1I8Q3Q7 A0A182QM56 A0A1A9YJZ2 E9HNI4 A0A0P5VB21 B7QEP7 A0A0P5TJ22 A0A0P5V4Y1 A0A0L0BZU4 A0A0P6ELN4 A0A0P5W1G6 A0A0P5YED8 A0A1J1HWC7 A0A0P5VBX9 A0A0P6ER17 A0A0P5LPD9 A0A0P6ERF1 A0A0P5W1J3
A0A2J7RGI8 A0A2J7RGG8 A0A1B6F841 A0A0C9RLQ9 E2AFM9 E2BG87 A0A158NEL6 A0A310SCX5 A0A154PK27 A0A2A3EC09 A0A0L7REY6 A0A1W4WR92 A0A0N0BKX5 A0A087ZXM1 A0A026WUN4 E0VUX7 E9IJB0 A0A0C9QD72 A0A195FLJ9 K7IYF5 A0A151XJ50 A0A232F2J8 A0A195BUW4 F4WRU4 A0A195BZW8 A0A195EN35 A0A1B6JEH0 A0A1B0D8A5 A0A2P8Y5Z3 A0A1B0CLA7 A0A336MX73 A0A336MS44 Q16N82 A0A3L8DGG3 A0A1Q3F0S6 A0A0J7KJK2 B0WR47 A0A182H4U1 A0A182R7L4 A0A084W9G5 A0A182MRE3 A0A182PEI2 A0A182W2L4 A0A182JID5 A0A182NG84 A0A182XLY0 Q7PTK3 A0A182HS30 A0A182XVD6 N6UCT2 A0A182US67 A0A182TSC2 A0A146LYB8 A0A0A9WSZ0 A0A182TX77 A0A0K8SVE8 T1J846 A0A224YNK6 A0A182FPZ3 D3TPL8 A0A1A9Z9U0 A0A2S2R0W1 A0A1B0FCI7 A0A1A9VG93 A0A131YFF8 A0A1E1XBT0 A0A131XC71 W5JUQ4 A0A2R5LJL0 A0A293MD54 J9K7L3 A0A2H8TFN9 A0A1B0BW91 A0A087T509 A0A1I8Q3Q7 A0A182QM56 A0A1A9YJZ2 E9HNI4 A0A0P5VB21 B7QEP7 A0A0P5TJ22 A0A0P5V4Y1 A0A0L0BZU4 A0A0P6ELN4 A0A0P5W1G6 A0A0P5YED8 A0A1J1HWC7 A0A0P5VBX9 A0A0P6ER17 A0A0P5LPD9 A0A0P6ERF1 A0A0P5W1J3
Pubmed
26354079
28004739
18362917
19820115
20798317
21347285
+ More
24508170 20566863 21282665 20075255 28648823 21719571 29403074 17510324 30249741 26483478 24438588 12364791 14747013 17210077 25244985 23537049 26823975 25401762 28797301 20353571 26830274 28503490 28049606 20920257 23761445 21292972 26108605
24508170 20566863 21282665 20075255 28648823 21719571 29403074 17510324 30249741 26483478 24438588 12364791 14747013 17210077 25244985 23537049 26823975 25401762 28797301 20353571 26830274 28503490 28049606 20920257 23761445 21292972 26108605
EMBL
KQ459232
KPJ02727.1
KQ461155
KPJ08207.1
NWSH01000075
PCG79915.1
+ More
GEZM01093063 JAV56365.1 KQ971361 EFA08805.1 NEVH01003784 PNF39938.1 PNF39935.1 GECZ01023392 JAS46377.1 GBYB01007916 JAG77683.1 GL439118 EFN67780.1 GL448116 EFN85295.1 ADTU01013497 KQ763581 OAD54917.1 KQ434931 KZC11814.1 KZ288288 PBC29313.1 KQ414608 KOC69390.1 KQ435689 KOX81211.1 KK107109 EZA59376.1 DS235797 EEB17183.1 GL763764 EFZ19321.1 GBYB01001254 JAG71021.1 KQ981490 KYN41211.1 KQ982080 KYQ60308.1 NNAY01001206 OXU24739.1 KQ976401 KYM92372.1 GL888292 EGI63101.1 KQ978501 KYM93453.1 KQ978625 KYN29581.1 GECU01010198 JAS97508.1 AJVK01004015 PYGN01000890 PSN39663.1 AJWK01017063 UFQS01002181 UFQT01002181 SSX13436.1 SSX32867.1 UFQT01002195 SSX32900.1 CH477835 EAT35794.1 QOIP01000008 RLU19530.1 GFDL01013894 JAV21151.1 LBMM01006664 KMQ90411.1 DS232051 EDS33153.1 JXUM01025646 KQ560729 KXJ81113.1 ATLV01021759 KE525322 KFB46859.1 AXCM01000365 AAAB01008799 EAA03791.5 APCN01000234 APGK01039907 APGK01039908 KB740975 KB632186 ENN76457.1 ERL89743.1 GDHC01006967 JAQ11662.1 GBHO01033068 GBHO01020674 JAG10536.1 JAG22930.1 GBRD01008700 JAG57121.1 JH431948 GFPF01004677 MAA15823.1 EZ423370 ADD19646.1 GGMS01013809 MBY83012.1 CCAG010011485 GEDV01010493 JAP78064.1 GFAC01002471 JAT96717.1 GEFH01004911 JAP63670.1 ADMH02000315 ETN67008.1 GGLE01005606 MBY09732.1 GFWV01013282 MAA38011.1 ABLF02038160 GFXV01001111 MBW12916.1 JXJN01021666 KK113438 KFM60198.1 AXCN02001629 GL732696 EFX66708.1 GDIP01102435 JAM01280.1 ABJB011045320 DS921995 EEC17319.1 GDIP01125588 GDIP01109989 LRGB01002860 JAL78126.1 KZS05824.1 GDIP01105291 JAL98423.1 JRES01001097 KNC25575.1 GDIQ01061898 JAN32839.1 GDIP01092284 JAM11431.1 GDIP01060789 JAM42926.1 CVRI01000027 CRK92407.1 GDIP01102434 JAM01281.1 GDIQ01072506 JAN22231.1 GDIQ01177361 JAK74364.1 GDIQ01072505 JAN22232.1 GDIP01092285 JAM11430.1
GEZM01093063 JAV56365.1 KQ971361 EFA08805.1 NEVH01003784 PNF39938.1 PNF39935.1 GECZ01023392 JAS46377.1 GBYB01007916 JAG77683.1 GL439118 EFN67780.1 GL448116 EFN85295.1 ADTU01013497 KQ763581 OAD54917.1 KQ434931 KZC11814.1 KZ288288 PBC29313.1 KQ414608 KOC69390.1 KQ435689 KOX81211.1 KK107109 EZA59376.1 DS235797 EEB17183.1 GL763764 EFZ19321.1 GBYB01001254 JAG71021.1 KQ981490 KYN41211.1 KQ982080 KYQ60308.1 NNAY01001206 OXU24739.1 KQ976401 KYM92372.1 GL888292 EGI63101.1 KQ978501 KYM93453.1 KQ978625 KYN29581.1 GECU01010198 JAS97508.1 AJVK01004015 PYGN01000890 PSN39663.1 AJWK01017063 UFQS01002181 UFQT01002181 SSX13436.1 SSX32867.1 UFQT01002195 SSX32900.1 CH477835 EAT35794.1 QOIP01000008 RLU19530.1 GFDL01013894 JAV21151.1 LBMM01006664 KMQ90411.1 DS232051 EDS33153.1 JXUM01025646 KQ560729 KXJ81113.1 ATLV01021759 KE525322 KFB46859.1 AXCM01000365 AAAB01008799 EAA03791.5 APCN01000234 APGK01039907 APGK01039908 KB740975 KB632186 ENN76457.1 ERL89743.1 GDHC01006967 JAQ11662.1 GBHO01033068 GBHO01020674 JAG10536.1 JAG22930.1 GBRD01008700 JAG57121.1 JH431948 GFPF01004677 MAA15823.1 EZ423370 ADD19646.1 GGMS01013809 MBY83012.1 CCAG010011485 GEDV01010493 JAP78064.1 GFAC01002471 JAT96717.1 GEFH01004911 JAP63670.1 ADMH02000315 ETN67008.1 GGLE01005606 MBY09732.1 GFWV01013282 MAA38011.1 ABLF02038160 GFXV01001111 MBW12916.1 JXJN01021666 KK113438 KFM60198.1 AXCN02001629 GL732696 EFX66708.1 GDIP01102435 JAM01280.1 ABJB011045320 DS921995 EEC17319.1 GDIP01125588 GDIP01109989 LRGB01002860 JAL78126.1 KZS05824.1 GDIP01105291 JAL98423.1 JRES01001097 KNC25575.1 GDIQ01061898 JAN32839.1 GDIP01092284 JAM11431.1 GDIP01060789 JAM42926.1 CVRI01000027 CRK92407.1 GDIP01102434 JAM01281.1 GDIQ01072506 JAN22231.1 GDIQ01177361 JAK74364.1 GDIQ01072505 JAN22232.1 GDIP01092285 JAM11430.1
Proteomes
UP000053268
UP000053240
UP000218220
UP000192223
UP000007266
UP000235965
+ More
UP000000311 UP000008237 UP000005205 UP000076502 UP000242457 UP000053825 UP000053105 UP000005203 UP000053097 UP000009046 UP000078541 UP000002358 UP000075809 UP000215335 UP000078540 UP000007755 UP000078542 UP000078492 UP000092462 UP000245037 UP000092461 UP000008820 UP000279307 UP000036403 UP000002320 UP000069940 UP000249989 UP000075900 UP000030765 UP000075883 UP000075885 UP000075920 UP000075880 UP000075884 UP000076407 UP000007062 UP000075840 UP000076408 UP000019118 UP000030742 UP000075903 UP000075902 UP000069272 UP000092445 UP000092444 UP000078200 UP000000673 UP000007819 UP000092460 UP000054359 UP000095300 UP000075886 UP000092443 UP000000305 UP000001555 UP000076858 UP000037069 UP000183832
UP000000311 UP000008237 UP000005205 UP000076502 UP000242457 UP000053825 UP000053105 UP000005203 UP000053097 UP000009046 UP000078541 UP000002358 UP000075809 UP000215335 UP000078540 UP000007755 UP000078542 UP000078492 UP000092462 UP000245037 UP000092461 UP000008820 UP000279307 UP000036403 UP000002320 UP000069940 UP000249989 UP000075900 UP000030765 UP000075883 UP000075885 UP000075920 UP000075880 UP000075884 UP000076407 UP000007062 UP000075840 UP000076408 UP000019118 UP000030742 UP000075903 UP000075902 UP000069272 UP000092445 UP000092444 UP000078200 UP000000673 UP000007819 UP000092460 UP000054359 UP000095300 UP000075886 UP000092443 UP000000305 UP000001555 UP000076858 UP000037069 UP000183832
Pfam
PF10221 DUF2151
Interpro
IPR019355
Cell_cycle_regulator_Mat89Bb
ProteinModelPortal
A0A194QB89
A0A194QRU2
A0A2A4K6L8
A0A1W4X207
A0A1Y1K9K7
D6WX84
+ More
A0A2J7RGI8 A0A2J7RGG8 A0A1B6F841 A0A0C9RLQ9 E2AFM9 E2BG87 A0A158NEL6 A0A310SCX5 A0A154PK27 A0A2A3EC09 A0A0L7REY6 A0A1W4WR92 A0A0N0BKX5 A0A087ZXM1 A0A026WUN4 E0VUX7 E9IJB0 A0A0C9QD72 A0A195FLJ9 K7IYF5 A0A151XJ50 A0A232F2J8 A0A195BUW4 F4WRU4 A0A195BZW8 A0A195EN35 A0A1B6JEH0 A0A1B0D8A5 A0A2P8Y5Z3 A0A1B0CLA7 A0A336MX73 A0A336MS44 Q16N82 A0A3L8DGG3 A0A1Q3F0S6 A0A0J7KJK2 B0WR47 A0A182H4U1 A0A182R7L4 A0A084W9G5 A0A182MRE3 A0A182PEI2 A0A182W2L4 A0A182JID5 A0A182NG84 A0A182XLY0 Q7PTK3 A0A182HS30 A0A182XVD6 N6UCT2 A0A182US67 A0A182TSC2 A0A146LYB8 A0A0A9WSZ0 A0A182TX77 A0A0K8SVE8 T1J846 A0A224YNK6 A0A182FPZ3 D3TPL8 A0A1A9Z9U0 A0A2S2R0W1 A0A1B0FCI7 A0A1A9VG93 A0A131YFF8 A0A1E1XBT0 A0A131XC71 W5JUQ4 A0A2R5LJL0 A0A293MD54 J9K7L3 A0A2H8TFN9 A0A1B0BW91 A0A087T509 A0A1I8Q3Q7 A0A182QM56 A0A1A9YJZ2 E9HNI4 A0A0P5VB21 B7QEP7 A0A0P5TJ22 A0A0P5V4Y1 A0A0L0BZU4 A0A0P6ELN4 A0A0P5W1G6 A0A0P5YED8 A0A1J1HWC7 A0A0P5VBX9 A0A0P6ER17 A0A0P5LPD9 A0A0P6ERF1 A0A0P5W1J3
A0A2J7RGI8 A0A2J7RGG8 A0A1B6F841 A0A0C9RLQ9 E2AFM9 E2BG87 A0A158NEL6 A0A310SCX5 A0A154PK27 A0A2A3EC09 A0A0L7REY6 A0A1W4WR92 A0A0N0BKX5 A0A087ZXM1 A0A026WUN4 E0VUX7 E9IJB0 A0A0C9QD72 A0A195FLJ9 K7IYF5 A0A151XJ50 A0A232F2J8 A0A195BUW4 F4WRU4 A0A195BZW8 A0A195EN35 A0A1B6JEH0 A0A1B0D8A5 A0A2P8Y5Z3 A0A1B0CLA7 A0A336MX73 A0A336MS44 Q16N82 A0A3L8DGG3 A0A1Q3F0S6 A0A0J7KJK2 B0WR47 A0A182H4U1 A0A182R7L4 A0A084W9G5 A0A182MRE3 A0A182PEI2 A0A182W2L4 A0A182JID5 A0A182NG84 A0A182XLY0 Q7PTK3 A0A182HS30 A0A182XVD6 N6UCT2 A0A182US67 A0A182TSC2 A0A146LYB8 A0A0A9WSZ0 A0A182TX77 A0A0K8SVE8 T1J846 A0A224YNK6 A0A182FPZ3 D3TPL8 A0A1A9Z9U0 A0A2S2R0W1 A0A1B0FCI7 A0A1A9VG93 A0A131YFF8 A0A1E1XBT0 A0A131XC71 W5JUQ4 A0A2R5LJL0 A0A293MD54 J9K7L3 A0A2H8TFN9 A0A1B0BW91 A0A087T509 A0A1I8Q3Q7 A0A182QM56 A0A1A9YJZ2 E9HNI4 A0A0P5VB21 B7QEP7 A0A0P5TJ22 A0A0P5V4Y1 A0A0L0BZU4 A0A0P6ELN4 A0A0P5W1G6 A0A0P5YED8 A0A1J1HWC7 A0A0P5VBX9 A0A0P6ER17 A0A0P5LPD9 A0A0P6ERF1 A0A0P5W1J3
Ontologies
GO
PANTHER
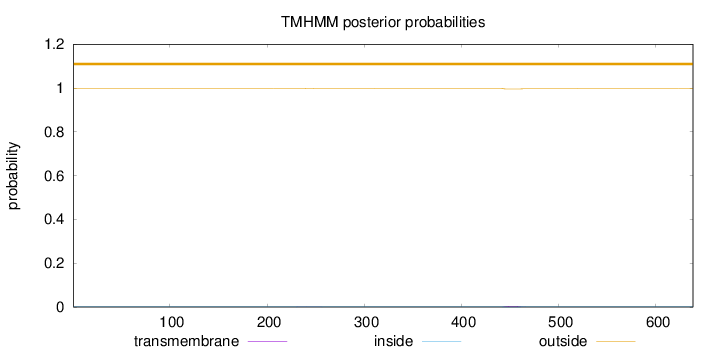
Topology
Length:
638
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08711
Exp number, first 60 AAs:
0.00157
Total prob of N-in:
0.00113
outside
1 - 638
Population Genetic Test Statistics
Pi
231.288957
Theta
162.7095
Tajima's D
1.753724
CLR
0.005679
CSRT
0.83850807459627
Interpretation
Uncertain
