Gene
KWMTBOMO10243 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA007315
Annotation
PREDICTED:_ubiquitin_carboxyl-terminal_hydrolase_7-like_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.509 Nuclear Reliability : 1.917
Sequence
CDS
ATGAATCACACGCCTGCACCTGACAGGCAGCAGCACGACTCCAATGTTAACCAGGTCGTGGAGATGGAGACTCAGGACGTTGAAACAATTACCACTGAAAAAACCTGGAATGAAGAAATGATGAAGGGTCCTAATGGTGAGGTTGTCATGGCTGATGTTATGAAGACACAGGAGACTGCCTCAGCTGAGATGCCCCTCGCCTGTCTTGACGCTGAAATGGAAGACGACGAAGCGCGTTCAGAGGCAACGTTCCGATTCACGGTCCACAACTTTAAGAATTTGAAAGACTCGATGCTGTCGCCGCCGTGCTACGTGCGCAATCTGCCGTGGAAGATAATGGTTATGCCGCGGCAAGCGCCTTCGCCCGATCGGCAACAGCAAAAGTCTCTGGGCTTCTTCCTACAGTGCAACGGGGAAAGTGAATCATCCAGCTGGTCTTGTTATGCGATGGCGGAACTGAGACTCATATCCCACAAGCCGGACACCGAACCTTTCCTAAGGAAAATTCAACATTTATTTTACAGCAAAGAAAACGACTGGGGCTTTTCTCACTTCATGACGTGGAACGACGTGTTAGACCCCGAGAAGGGCTACATTAAAGACGACGCGATCACTCTGGAAGTTCACGTCACCGCCGAAGCACCCCACGGCGTATCTTGGGATTCTAAGAAGCATACAGGATATGTTGGTCTGAAGAATCAAGGTGCAACTTGTTATATGAACTCGTTACTTCAAACACTTTATTTCACGAATCAACTCCGCAAAGCTGTGTACAAGATGCCTACAGAATCGGACGATAGTACTAGATCGGTGGCTTTGGCTTTGCAAAGAGTATTCTACGAGTTACAGTTCTGTGATAAACCAGTTGGTACAAAGAAGTTGACCAAGAGCTTTGGTTGGGAAACCCTTGATTCTTTCATGCAACACGACGTACAAGAGTTCCTTAGGGTTTTATTGGATAAATTAGAAAGTAAAATGAAGGGCACATGCGTGGAGGGCACGGTTCCTCGACTGTTCGAGGGCAAGATGACATCATACATTAAGTGTAAGAACGTTAACGTTTCAAGTACCCGAGTGGAAACATTCTACGATATCCAACTCAATATAAAAGGGAAGAAAAATATCTACGAATCGTTCAAAGATTACATAAGCACAGAAACGCTAGACGGCGAGAATAAGTACGACGCTGGCGAGCACGGTCTCCAGGAAGCCGAGAAAGGCGTCATATTTGCCACGTTCCCGCCCGTGTTACATTTACACTTGATGAGATTCCAATACGATCCCATTACCGACAGTTCGGTCAAGTTTAATGACAGATTTGAATTCTACGAACGTATCAATTTAGATGCATATCTACAGGAGAAACCAGAAACACCGGCTGACTACACCCTCCATGCGGTATTAGTTCACTCGGGAGACAACCACGGGGGGCATTATGTGGTCTTTATCAATCCGAAAGGTGACGGAAAGTGGTGTAAGTTCGACGACGACGTGGTGTCCCGGTGCACGAAACAGGAGGCCATCGAATACAACTACGGCGGTCACGACGAGGACATGACGTTGACCGTTCGCCACTGCACTAACGCCTATATGTTGGTTTATATCAGGGATTCACAGTTGAAGACTGTTCTACAAGAAGTAACACAAGCAGACATACCTACTGAGTTGAGTGAGAGATTAGCTGAAGAGAAGAGAATCGAAACCATTCGTCGCAAAGAGCGGAACGAAGCCCACCTGTATATGAACGTGAATGTAGTATTGGAGGAAGCTTTCGACGGTCATCAAGGGAACGATCTGTACGACCCGGAGCGCGCGCACTACCGCGTGTTCCGCGTCAGGAAGCAGGCCACCGTCTCTGAACTCATGGAGATATTGTCAGAGAACTTTAAGTATCCACTGCAGCAGCTCCGACCCTGGCCATTTAGCGCACGCTCTAATCAGACATGCAGACCGACCTGCCTAGACATTTTTAACGATCAACTCAAGACTGTAGCTGACGTTTCTGAGAACATGAACCCATGGAACATATTCCTCGAGATGCTGCCTCCAGATTCTGGTCTCACATCATTGCCTTGGTTCGATAAGAAGAACGACGTGGTCCTGTTTTTTAAGTATTATGATCCTAAACAGAAGCGCATACACTACTGCGGACACCATTATTTGCCCATCGCAAGCAAACCTGCCGATCTTCTACCTATACTCAATCAACGTGCAGGATTCCCACCAGACACTCCACTAGTTCTGTATGAAGAAATCAAACCAGACTTTGTTGAAAAAATCAGTAATTACAACGAACCTCTAGAAAAGGTATTGGACGAGCTGATGGACGGCGACATCATTGTGTTCGAACGTGCCGACAACCGACACGACGAGCTGGAGCTGCCCACCTGCCAGGACTACTTCAAGTACATCTTCTACAAGGTCGAGGTGCAGTTCGTCGATAAAACGGTGCCCAATGATCCGGGATTCACAATGGAGCTATCGATGCAAATGCGTTATGATCAGATGGCGCGGGCGGTCGGCCAGAGGCTCAACGTGGACCCTTACTTAATACAGTTCTTTAAGTGTCAAAACTACAAGGACACCCCAGGACTACCTTTGAGATACTCATACGACGGAATACTTAAGGACTTATTAGTGTACTGCAAACCAAAATGCCCTAAGAAATTATTCTATCAGATATTATCTATTAAAGTAAACGAATTGGACAACAAGAAGCAATTCAAATGTCTATGGGTTGGTCCAAATTACAAAGAGGATAAAGAATTAATTTTGTATCCAAATAAAGGTGGTAAAGTGTCGGACATTTTAGAAGAGGCAGCTAAAGTAGTCGAGATGTCAACCGAGGGCTCAGGCAGGCTAAGGATCGTGGAGGTGTCGTGTCACAAAGTGCTACCGGGTCCGGACCCCGAGCTCACACTCGACCAGGTCACCATATCGCCGCCCAGACTCTACAGAATAGAGGAGATACCTAGAGACGAACTGAATTTACAAGAGGATGAGATGCTGGTTCCGTGTGCGCATTTCTACAAGCAAGTATACGCGACGTTCGGAATTCCGTTTTACGCGCGTTTCAAACACAACGAACCGTTCGAAGCTGTCAAAGATCGCCTACAGAAGAAACTCGACATACCCGACAAAGAATGGGAAAAGTACAACTTTGCTGTAGTGACAAACGGCAGACCTAATTACATAAGCGAAGGAGCGACAGTGAATATCAACGATTTCAGACTAAGTAGTAATACGAATCTGTCGCACCCAGACGCAACGGCGCGGCCCTGGTTCGGGCTCGAGCACATCAACAAGACGCCGAAGCGATCCCGCGTCAACTACTTGGAGAAGGCCATCAAAATATACAACTGA
Protein
MNHTPAPDRQQHDSNVNQVVEMETQDVETITTEKTWNEEMMKGPNGEVVMADVMKTQETASAEMPLACLDAEMEDDEARSEATFRFTVHNFKNLKDSMLSPPCYVRNLPWKIMVMPRQAPSPDRQQQKSLGFFLQCNGESESSSWSCYAMAELRLISHKPDTEPFLRKIQHLFYSKENDWGFSHFMTWNDVLDPEKGYIKDDAITLEVHVTAEAPHGVSWDSKKHTGYVGLKNQGATCYMNSLLQTLYFTNQLRKAVYKMPTESDDSTRSVALALQRVFYELQFCDKPVGTKKLTKSFGWETLDSFMQHDVQEFLRVLLDKLESKMKGTCVEGTVPRLFEGKMTSYIKCKNVNVSSTRVETFYDIQLNIKGKKNIYESFKDYISTETLDGENKYDAGEHGLQEAEKGVIFATFPPVLHLHLMRFQYDPITDSSVKFNDRFEFYERINLDAYLQEKPETPADYTLHAVLVHSGDNHGGHYVVFINPKGDGKWCKFDDDVVSRCTKQEAIEYNYGGHDEDMTLTVRHCTNAYMLVYIRDSQLKTVLQEVTQADIPTELSERLAEEKRIETIRRKERNEAHLYMNVNVVLEEAFDGHQGNDLYDPERAHYRVFRVRKQATVSELMEILSENFKYPLQQLRPWPFSARSNQTCRPTCLDIFNDQLKTVADVSENMNPWNIFLEMLPPDSGLTSLPWFDKKNDVVLFFKYYDPKQKRIHYCGHHYLPIASKPADLLPILNQRAGFPPDTPLVLYEEIKPDFVEKISNYNEPLEKVLDELMDGDIIVFERADNRHDELELPTCQDYFKYIFYKVEVQFVDKTVPNDPGFTMELSMQMRYDQMARAVGQRLNVDPYLIQFFKCQNYKDTPGLPLRYSYDGILKDLLVYCKPKCPKKLFYQILSIKVNELDNKKQFKCLWVGPNYKEDKELILYPNKGGKVSDILEEAAKVVEMSTEGSGRLRIVEVSCHKVLPGPDPELTLDQVTISPPRLYRIEEIPRDELNLQEDEMLVPCAHFYKQVYATFGIPFYARFKHNEPFEAVKDRLQKKLDIPDKEWEKYNFAVVTNGRPNYISEGATVNINDFRLSSNTNLSHPDATARPWFGLEHINKTPKRSRVNYLEKAIKIYN
Summary
Similarity
Belongs to the peptidase C19 family.
Uniprot
H9JCR8
A0A1E1W4F0
A0A2A4K8B3
A0A2A4K755
A0A2H1WIL8
A0A1E1WT07
+ More
A0A2A4K6P7 A0A2A4K802 A0A2A4K8B9 A0A2A4K7M5 A0A194QH05 A0A212F6V4 A0A2J7PLX9 A0A1W4XGG6 A0A1Y1NCM6 A0A1W4XR12 A0A067RK91 A0A1W4XRW7 D6WV88 A0A1W4XGU8 A0A3L8DDC9 A0A151XHI5 A0A158P2W3 A0A195DFJ6 A0A0T6B0P9 F4W5Y2 A0A195CY21 E2BIS3 A0A195EZU6 A0A026WG09 A0A0C9RA19 V9I9T1 A0A154PQZ5 A0A2P8XES8 A0A1B6MRK8 A0A195BN65 E0VQ72 A0A0C9R494 A0A069DZQ5 A0A146M5G6 A0A3R7ML36 N6U732 A0A023F463 A0A146KTM6 A0A224X5V0 A0A146KS22 T1J1R9 V5HKQ0 A0A1E1XR42 A0A2R5LCL6 A0A224Z9C1 A0A293MIZ3 U4U9K9 A0A023GFZ8 A0A131YJQ5 A0A1E1X577 A0A1L8DTL1 A0A131YLU4 A0A384WIE7 A0A087U529 A0A3S3PLW8 A0A2C9JFV6 A0A2L2XYY8 A0A2L2XZ45 A0A210QGU4 A0A3S1B020 A0A2P2I3R0 A0A1S3HQU7 K1P6N8 T1KCD8 A0A0K8TFR6 A0A0P5JWR9 A0A0P5NQS4 A0A0P5K081 A0A0P5NWX5 A0A0P6DJJ2 A0A0N8E5Z1 A0A0P5L5A0 A0A0P5HMY0 E9HA21 A0A0P5Z0B1 A0A0K8TFR1 A0A0P6IE42 A0A0P5IUI7 A0A0P6FHF0 A0A0P5K914 A0A0P5N2Y7 A0A0P5SIW8 V3ZII5 A0A0N8CCP8 A0A218UFY2 A0A151N170 A0A2I0U2E9 G1N8T7 R7TEG5 U3JN08 A0A099Z3M7 A0A1W5AAR5 A0A1D5PPD3
A0A2A4K6P7 A0A2A4K802 A0A2A4K8B9 A0A2A4K7M5 A0A194QH05 A0A212F6V4 A0A2J7PLX9 A0A1W4XGG6 A0A1Y1NCM6 A0A1W4XR12 A0A067RK91 A0A1W4XRW7 D6WV88 A0A1W4XGU8 A0A3L8DDC9 A0A151XHI5 A0A158P2W3 A0A195DFJ6 A0A0T6B0P9 F4W5Y2 A0A195CY21 E2BIS3 A0A195EZU6 A0A026WG09 A0A0C9RA19 V9I9T1 A0A154PQZ5 A0A2P8XES8 A0A1B6MRK8 A0A195BN65 E0VQ72 A0A0C9R494 A0A069DZQ5 A0A146M5G6 A0A3R7ML36 N6U732 A0A023F463 A0A146KTM6 A0A224X5V0 A0A146KS22 T1J1R9 V5HKQ0 A0A1E1XR42 A0A2R5LCL6 A0A224Z9C1 A0A293MIZ3 U4U9K9 A0A023GFZ8 A0A131YJQ5 A0A1E1X577 A0A1L8DTL1 A0A131YLU4 A0A384WIE7 A0A087U529 A0A3S3PLW8 A0A2C9JFV6 A0A2L2XYY8 A0A2L2XZ45 A0A210QGU4 A0A3S1B020 A0A2P2I3R0 A0A1S3HQU7 K1P6N8 T1KCD8 A0A0K8TFR6 A0A0P5JWR9 A0A0P5NQS4 A0A0P5K081 A0A0P5NWX5 A0A0P6DJJ2 A0A0N8E5Z1 A0A0P5L5A0 A0A0P5HMY0 E9HA21 A0A0P5Z0B1 A0A0K8TFR1 A0A0P6IE42 A0A0P5IUI7 A0A0P6FHF0 A0A0P5K914 A0A0P5N2Y7 A0A0P5SIW8 V3ZII5 A0A0N8CCP8 A0A218UFY2 A0A151N170 A0A2I0U2E9 G1N8T7 R7TEG5 U3JN08 A0A099Z3M7 A0A1W5AAR5 A0A1D5PPD3
Pubmed
EMBL
BABH01041594
BABH01041595
GDQN01009199
JAT81855.1
NWSH01000075
PCG79910.1
+ More
PCG79909.1 ODYU01008918 SOQ52915.1 GDQN01000950 JAT90104.1 PCG79911.1 PCG79913.1 PCG79912.1 PCG79914.1 KQ459232 KPJ02726.1 AGBW02009963 OWR49471.1 NEVH01024423 PNF17329.1 GEZM01006600 JAV95692.1 KK852421 KDR24281.1 KQ971363 EFA07762.1 QOIP01000009 RLU18475.1 KQ982130 KYQ59748.1 ADTU01007519 ADTU01007520 ADTU01007521 ADTU01007522 KQ980903 KYN11665.1 LJIG01016292 KRT81039.1 GL887695 EGI70458.1 KQ977185 KYN05049.1 GL448530 EFN84413.1 KQ981905 KYN33399.1 KK107231 EZA55055.1 GBYB01009842 JAG79609.1 JR037624 AEY57878.1 KQ435012 KZC13774.1 PYGN01002425 PSN30510.1 GEBQ01001431 JAT38546.1 KQ976432 KYM87470.1 DS235392 EEB15528.1 GBYB01001636 JAG71403.1 GBGD01000230 JAC88659.1 GDHC01004633 JAQ13996.1 QCYY01001217 ROT79625.1 APGK01039902 APGK01039903 APGK01039904 KB740975 ENN76456.1 GBBI01002923 JAC15789.1 GDHC01018785 JAP99843.1 GFTR01008569 JAW07857.1 GDHC01019830 JAP98798.1 JH431789 GANP01010425 JAB74043.1 GFAA01001898 JAU01537.1 GGLE01002971 MBY07097.1 GFPF01013075 MAA24221.1 GFWV01020929 MAA45657.1 KB632186 ERL89742.1 GBBM01003510 JAC31908.1 GEDV01009033 JAP79524.1 GFAC01004785 JAT94403.1 GFDF01004459 JAV09625.1 GEDV01009032 JAP79525.1 MF680835 ATB19369.1 KK118215 KFM72468.1 NCKU01005814 RWS04141.1 IAAA01008906 LAA01156.1 IAAA01008907 LAA01158.1 NEDP02003741 OWF47975.1 RQTK01000653 RUS76556.1 IACF01003031 LAB68662.1 JH818502 EKC19292.1 CAEY01001959 GBRD01005931 GBRD01005930 GBRD01001389 JAG64432.1 GDIQ01193001 JAK58724.1 GDIQ01222984 GDIQ01139110 GDIQ01094828 GDIQ01031460 JAL12616.1 GDIQ01196596 JAK55129.1 GDIQ01136614 JAL15112.1 GDIQ01077477 JAN17260.1 GDIQ01058540 JAN36197.1 GDIQ01174894 JAK76831.1 GDIQ01231079 JAK20646.1 GL732610 EFX71348.1 GDIP01050837 JAM52878.1 GBRD01008013 GBRD01001394 GBRD01001393 GBRD01001392 GBRD01001391 GBRD01001390 JAG64427.1 GDIQ01005867 JAN88870.1 GDIQ01210334 JAK41391.1 GDIQ01094829 GDIQ01049455 JAN45282.1 GDIQ01193002 JAK58723.1 GDIQ01152014 JAK99711.1 GDIQ01092677 JAL59049.1 KB203566 ESO84027.1 GDIQ01092675 JAL59051.1 MUZQ01000332 OWK52645.1 AKHW03004154 KYO30488.1 KZ506306 PKU40254.1 AMQN01014839 KB311344 ELT89456.1 AGTO01020842 KL888978 KGL76312.1 AADN05000376
PCG79909.1 ODYU01008918 SOQ52915.1 GDQN01000950 JAT90104.1 PCG79911.1 PCG79913.1 PCG79912.1 PCG79914.1 KQ459232 KPJ02726.1 AGBW02009963 OWR49471.1 NEVH01024423 PNF17329.1 GEZM01006600 JAV95692.1 KK852421 KDR24281.1 KQ971363 EFA07762.1 QOIP01000009 RLU18475.1 KQ982130 KYQ59748.1 ADTU01007519 ADTU01007520 ADTU01007521 ADTU01007522 KQ980903 KYN11665.1 LJIG01016292 KRT81039.1 GL887695 EGI70458.1 KQ977185 KYN05049.1 GL448530 EFN84413.1 KQ981905 KYN33399.1 KK107231 EZA55055.1 GBYB01009842 JAG79609.1 JR037624 AEY57878.1 KQ435012 KZC13774.1 PYGN01002425 PSN30510.1 GEBQ01001431 JAT38546.1 KQ976432 KYM87470.1 DS235392 EEB15528.1 GBYB01001636 JAG71403.1 GBGD01000230 JAC88659.1 GDHC01004633 JAQ13996.1 QCYY01001217 ROT79625.1 APGK01039902 APGK01039903 APGK01039904 KB740975 ENN76456.1 GBBI01002923 JAC15789.1 GDHC01018785 JAP99843.1 GFTR01008569 JAW07857.1 GDHC01019830 JAP98798.1 JH431789 GANP01010425 JAB74043.1 GFAA01001898 JAU01537.1 GGLE01002971 MBY07097.1 GFPF01013075 MAA24221.1 GFWV01020929 MAA45657.1 KB632186 ERL89742.1 GBBM01003510 JAC31908.1 GEDV01009033 JAP79524.1 GFAC01004785 JAT94403.1 GFDF01004459 JAV09625.1 GEDV01009032 JAP79525.1 MF680835 ATB19369.1 KK118215 KFM72468.1 NCKU01005814 RWS04141.1 IAAA01008906 LAA01156.1 IAAA01008907 LAA01158.1 NEDP02003741 OWF47975.1 RQTK01000653 RUS76556.1 IACF01003031 LAB68662.1 JH818502 EKC19292.1 CAEY01001959 GBRD01005931 GBRD01005930 GBRD01001389 JAG64432.1 GDIQ01193001 JAK58724.1 GDIQ01222984 GDIQ01139110 GDIQ01094828 GDIQ01031460 JAL12616.1 GDIQ01196596 JAK55129.1 GDIQ01136614 JAL15112.1 GDIQ01077477 JAN17260.1 GDIQ01058540 JAN36197.1 GDIQ01174894 JAK76831.1 GDIQ01231079 JAK20646.1 GL732610 EFX71348.1 GDIP01050837 JAM52878.1 GBRD01008013 GBRD01001394 GBRD01001393 GBRD01001392 GBRD01001391 GBRD01001390 JAG64427.1 GDIQ01005867 JAN88870.1 GDIQ01210334 JAK41391.1 GDIQ01094829 GDIQ01049455 JAN45282.1 GDIQ01193002 JAK58723.1 GDIQ01152014 JAK99711.1 GDIQ01092677 JAL59049.1 KB203566 ESO84027.1 GDIQ01092675 JAL59051.1 MUZQ01000332 OWK52645.1 AKHW03004154 KYO30488.1 KZ506306 PKU40254.1 AMQN01014839 KB311344 ELT89456.1 AGTO01020842 KL888978 KGL76312.1 AADN05000376
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000235965
UP000192223
+ More
UP000027135 UP000007266 UP000279307 UP000075809 UP000005205 UP000078492 UP000007755 UP000078542 UP000008237 UP000078541 UP000053097 UP000076502 UP000245037 UP000078540 UP000009046 UP000283509 UP000019118 UP000030742 UP000054359 UP000285301 UP000076420 UP000242188 UP000271974 UP000085678 UP000005408 UP000015104 UP000000305 UP000030746 UP000197619 UP000050525 UP000001645 UP000014760 UP000016665 UP000053641 UP000192224 UP000000539
UP000027135 UP000007266 UP000279307 UP000075809 UP000005205 UP000078492 UP000007755 UP000078542 UP000008237 UP000078541 UP000053097 UP000076502 UP000245037 UP000078540 UP000009046 UP000283509 UP000019118 UP000030742 UP000054359 UP000285301 UP000076420 UP000242188 UP000271974 UP000085678 UP000005408 UP000015104 UP000000305 UP000030746 UP000197619 UP000050525 UP000001645 UP000014760 UP000016665 UP000053641 UP000192224 UP000000539
Interpro
IPR018200
USP_CS
+ More
IPR038765 Papain-like_cys_pep_sf
IPR001394 Peptidase_C19_UCH
IPR002083 MATH/TRAF_dom
IPR028889 USP_dom
IPR008974 TRAF-like
IPR029346 USP_C
IPR024729 USP7_ICP0-binding_dom
IPR000300 IPPc
IPR036691 Endo/exonu/phosph_ase_sf
IPR002130 Cyclophilin-type_PPIase_dom
IPR029000 Cyclophilin-like_dom_sf
IPR038765 Papain-like_cys_pep_sf
IPR001394 Peptidase_C19_UCH
IPR002083 MATH/TRAF_dom
IPR028889 USP_dom
IPR008974 TRAF-like
IPR029346 USP_C
IPR024729 USP7_ICP0-binding_dom
IPR000300 IPPc
IPR036691 Endo/exonu/phosph_ase_sf
IPR002130 Cyclophilin-type_PPIase_dom
IPR029000 Cyclophilin-like_dom_sf
Gene 3D
ProteinModelPortal
H9JCR8
A0A1E1W4F0
A0A2A4K8B3
A0A2A4K755
A0A2H1WIL8
A0A1E1WT07
+ More
A0A2A4K6P7 A0A2A4K802 A0A2A4K8B9 A0A2A4K7M5 A0A194QH05 A0A212F6V4 A0A2J7PLX9 A0A1W4XGG6 A0A1Y1NCM6 A0A1W4XR12 A0A067RK91 A0A1W4XRW7 D6WV88 A0A1W4XGU8 A0A3L8DDC9 A0A151XHI5 A0A158P2W3 A0A195DFJ6 A0A0T6B0P9 F4W5Y2 A0A195CY21 E2BIS3 A0A195EZU6 A0A026WG09 A0A0C9RA19 V9I9T1 A0A154PQZ5 A0A2P8XES8 A0A1B6MRK8 A0A195BN65 E0VQ72 A0A0C9R494 A0A069DZQ5 A0A146M5G6 A0A3R7ML36 N6U732 A0A023F463 A0A146KTM6 A0A224X5V0 A0A146KS22 T1J1R9 V5HKQ0 A0A1E1XR42 A0A2R5LCL6 A0A224Z9C1 A0A293MIZ3 U4U9K9 A0A023GFZ8 A0A131YJQ5 A0A1E1X577 A0A1L8DTL1 A0A131YLU4 A0A384WIE7 A0A087U529 A0A3S3PLW8 A0A2C9JFV6 A0A2L2XYY8 A0A2L2XZ45 A0A210QGU4 A0A3S1B020 A0A2P2I3R0 A0A1S3HQU7 K1P6N8 T1KCD8 A0A0K8TFR6 A0A0P5JWR9 A0A0P5NQS4 A0A0P5K081 A0A0P5NWX5 A0A0P6DJJ2 A0A0N8E5Z1 A0A0P5L5A0 A0A0P5HMY0 E9HA21 A0A0P5Z0B1 A0A0K8TFR1 A0A0P6IE42 A0A0P5IUI7 A0A0P6FHF0 A0A0P5K914 A0A0P5N2Y7 A0A0P5SIW8 V3ZII5 A0A0N8CCP8 A0A218UFY2 A0A151N170 A0A2I0U2E9 G1N8T7 R7TEG5 U3JN08 A0A099Z3M7 A0A1W5AAR5 A0A1D5PPD3
A0A2A4K6P7 A0A2A4K802 A0A2A4K8B9 A0A2A4K7M5 A0A194QH05 A0A212F6V4 A0A2J7PLX9 A0A1W4XGG6 A0A1Y1NCM6 A0A1W4XR12 A0A067RK91 A0A1W4XRW7 D6WV88 A0A1W4XGU8 A0A3L8DDC9 A0A151XHI5 A0A158P2W3 A0A195DFJ6 A0A0T6B0P9 F4W5Y2 A0A195CY21 E2BIS3 A0A195EZU6 A0A026WG09 A0A0C9RA19 V9I9T1 A0A154PQZ5 A0A2P8XES8 A0A1B6MRK8 A0A195BN65 E0VQ72 A0A0C9R494 A0A069DZQ5 A0A146M5G6 A0A3R7ML36 N6U732 A0A023F463 A0A146KTM6 A0A224X5V0 A0A146KS22 T1J1R9 V5HKQ0 A0A1E1XR42 A0A2R5LCL6 A0A224Z9C1 A0A293MIZ3 U4U9K9 A0A023GFZ8 A0A131YJQ5 A0A1E1X577 A0A1L8DTL1 A0A131YLU4 A0A384WIE7 A0A087U529 A0A3S3PLW8 A0A2C9JFV6 A0A2L2XYY8 A0A2L2XZ45 A0A210QGU4 A0A3S1B020 A0A2P2I3R0 A0A1S3HQU7 K1P6N8 T1KCD8 A0A0K8TFR6 A0A0P5JWR9 A0A0P5NQS4 A0A0P5K081 A0A0P5NWX5 A0A0P6DJJ2 A0A0N8E5Z1 A0A0P5L5A0 A0A0P5HMY0 E9HA21 A0A0P5Z0B1 A0A0K8TFR1 A0A0P6IE42 A0A0P5IUI7 A0A0P6FHF0 A0A0P5K914 A0A0P5N2Y7 A0A0P5SIW8 V3ZII5 A0A0N8CCP8 A0A218UFY2 A0A151N170 A0A2I0U2E9 G1N8T7 R7TEG5 U3JN08 A0A099Z3M7 A0A1W5AAR5 A0A1D5PPD3
PDB
5FWI
E-value=0,
Score=2318
Ontologies
GO
GO:0036459
GO:0016579
GO:0006511
GO:0005634
GO:0005829
GO:0004843
GO:0051090
GO:0008134
GO:0031647
GO:0004197
GO:0016021
GO:0016787
GO:0003755
GO:0050821
GO:1990380
GO:0032088
GO:0002039
GO:0035520
GO:0008022
GO:0010216
GO:0032435
GO:1901537
GO:0006283
GO:0031625
GO:0042752
GO:0032991
GO:1904888
GO:0005515
GO:0006397
GO:0005524
GO:0004812
GO:0006418
GO:0003723
GO:0046872
GO:0003824
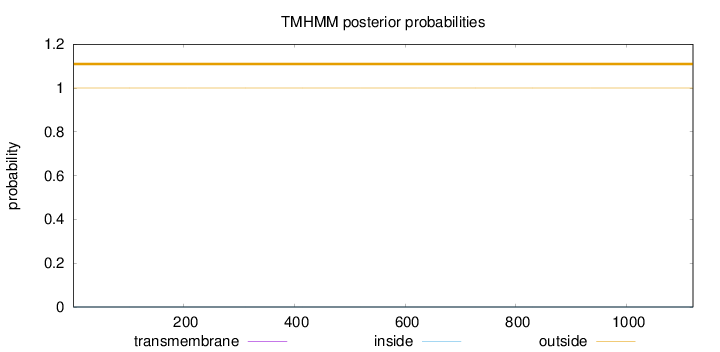
Topology
Length:
1120
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0037
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00009
outside
1 - 1120
Population Genetic Test Statistics
Pi
250.707418
Theta
186.648595
Tajima's D
1.011323
CLR
0.235401
CSRT
0.668366581670916
Interpretation
Uncertain
