Gene
KWMTBOMO10231
Pre Gene Modal
BGIBMGA000180
Annotation
PREDICTED:_serine/threonine-protein_kinase/endoribonuclease_IRE1_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.05 Nuclear Reliability : 1.072 PlasmaMembrane Reliability : 1.151
Sequence
CDS
ATGAAGTTTTTATTATTGATACTATTTCTGCTCGGTTCCTCCGGCTTCGGAACCGTTGTTGATGAAGAGGGAAAGGAAAGCACAGAAATATCCCGTGCTTTGGATGACCGGCCGCTTCTGTTTGCCACGTTAGGAGGCGGTATGGTTGCTGTTGATCCTGTGTCTGGAGACATTATTTGGAAATTAAAAGACGAGCCCGTTGTCAAAGTTCCGAGCCAAGACGCGAACTTGATGCCGCAGTTCCTCCCGGATCCGAGGCATGGATTCCTATACACGTACGGCCCGAGGGGCGACCAGCAAATGTTGAAGAAGCTCCCGTTTACAATACCGGAGCTGGTGGCAAACGCGCCCTGCCGATCTACAGACGGTATTTTGTACACCGGAAAGAAAAGCGATACTTGGTTCATGATTGATCCATTAACCGGCTTTCGTGAGCACGTTTCAGGTTTTGACAAATCGAAAATATTCAAAAGCGACGACGAGAACACTTGCAACTCCGAGAAGAAGAAAGGTATTTACGTGGCGCGTACTGAATACAACATCCTGATGCACGATTCGAAGAATGTAAATCATAAATGGAATGTTACGTTCTTTGATTACACGTCGCTAGCTATGGGCAAGGACATGTTGAATGATTACGGTACAATACATTTCACATCGACGTCGAACGGTCGCGTCATGTCTTTCGACCGGAAGACCGGCGACCTCGTCTGGTCCCACAACTTTGGGACTCCGGTCGTAGCTGCCTACCTCCTGGACAGAGACGGCCTTATCTCCGTCCCTTTCAACTCGATCGGTGACGACACGCTCGATCATATCATGGAGGACGCTAAAACCCTTTCGAACGGGAAAGGACTCAAAAATTCGAATATCGAATTGTACCCGACTTTATACGTCGGTGAGCATAACCACGGTCTATACGCGCTATCGTCGTTAGTCGATAAAAACACGATAACTATATCGACGGGTCATCAGCCCTTGCTCATCGAAGGCCCCTCGACGGAGACACTAAAGACGAACGCGCCGCGCTACGAGCCTATCAAGAACATCCACTACAAACTGAGCGACCTTAACTTGCACGTCTCGGCGCCGTATCTCCTTCTCGGCCACTACAAAGTTCCGGAACTGACGACCTCCTGGATACCCCATTTGCCTAACTCGAACGTTTTGAACCTGAGAGATCCACAAATCGATTCAATAAAACTAATCAACGGAGCAGTACAAGCTCAGACGGAGAATGACGTTGAGAACGAAACGAATTTGAAGTCCAATTCGGTATCAGTTTCGGTTCAGACTGAGGATTTGTTCGAGGGTTTGTCGTTCAGGCCCGATCTGTGGTACAAGCAGGCCCACGTTTGGCTCCACCAGCAGGAGAATAAAGTGCTGAAAGTGGCACTAATTATTCTCGTCGGTTTGGTCATCGCCATGTTCTGGTACTTAAGGCTGCAGGCTCGTGAGTTCCAACAACTATCGCAAGGCGGTTCCCGAAGCTCCACCTCATCTACGTCGTCCCAGGGCGAAGTTACCGGTCAGCTCATGGAAATAGGAGAGGGTGAAATGAAGATCGGAAAGATTACATTCCAGACAGATCAGGTGCTGGGAAAAGGTTGCGAAGGCACCTTCGTTTACAGAGGGACTTTCGATAAACGCGACGTGGCAGTAAAACGATTGCTCCCTGAGTGTTTCACGTTCGCCGACCGCGAGGTGGCGCTGCTGCGGGAGTCGGACGCGCACGCACACGTCGTGCGCTACTACTGTACTGAACGCGATAAGCAGTTCAGGTACATTGCACTAGAGCTATGTTCAGCGACATTGCAGGACTATGTTGAGAAGAAGTTGAACTTCGAGTGCAAGATTGATTCGGTCGAAATACTGAGGCAGGCCACTATGGGCTTATCGCACTTACACTCTATGGATATAGTACATAGGGATGTGAAGCCGCACAACGTTTTGCTGTCGATGCCGAGTGGGACCGGCGAGGTGCGCGCCATGATATCGGACTTCGGCCTGTCCAAGAAGCTGAACATAGGGCGGGTCAGCTTCTCGAGGCGCTCTGGGGTCACCGGCACCGACGGCTGGATAGCGCCGGAGATGATCAACGGCGAACGGACTACGACATCGATCGATATATTTTCGTTGGGCTGCGTGTTCTATTACGTATTGTCAAGAGGACAACATCCATTCGGTGATGTTTTGCGGCGTCAAGCGAACATACTCACCGGGGACTACAATTTGGATCACCTCGACAAAATTCTGCCTGAAGAGGAATTGGTAGTGGTTAAAATATTGATCCGAGCTATGATCTCGGCCAAGCCCAGCGCACGTCCTCCCTGCGAAACGATACTGAAATACCCGATGTTCTGGTGCAAGCAAAGCATTCTGAACTTCTTGCAGGACGTCAGCGATCAAGTGGAGAGTTGTGCGTGCGGTTTTGGGGCGGAGCACACGCTGGAGCGGGGCGGCGCGCGGGTGCTGCGCGGCGACTGGCGCGCGCGCGTGTGCCGCGTGGTGGCGGGCGACCTGCGCACGCGCCGCACGTACCGCGGGGACCGCGTCGCGCACCTGCTGCGGGCCATCCGGAACAAGGTACCGACACCGACCAGACCACATAGTGGTCTTCTTCTTCCGGCTGTTTTGAAGAGTATATACTGGGTGTTAGGATACCGCTTCCGAAAATGA
Protein
MKFLLLILFLLGSSGFGTVVDEEGKESTEISRALDDRPLLFATLGGGMVAVDPVSGDIIWKLKDEPVVKVPSQDANLMPQFLPDPRHGFLYTYGPRGDQQMLKKLPFTIPELVANAPCRSTDGILYTGKKSDTWFMIDPLTGFREHVSGFDKSKIFKSDDENTCNSEKKKGIYVARTEYNILMHDSKNVNHKWNVTFFDYTSLAMGKDMLNDYGTIHFTSTSNGRVMSFDRKTGDLVWSHNFGTPVVAAYLLDRDGLISVPFNSIGDDTLDHIMEDAKTLSNGKGLKNSNIELYPTLYVGEHNHGLYALSSLVDKNTITISTGHQPLLIEGPSTETLKTNAPRYEPIKNIHYKLSDLNLHVSAPYLLLGHYKVPELTTSWIPHLPNSNVLNLRDPQIDSIKLINGAVQAQTENDVENETNLKSNSVSVSVQTEDLFEGLSFRPDLWYKQAHVWLHQQENKVLKVALIILVGLVIAMFWYLRLQAREFQQLSQGGSRSSTSSTSSQGEVTGQLMEIGEGEMKIGKITFQTDQVLGKGCEGTFVYRGTFDKRDVAVKRLLPECFTFADREVALLRESDAHAHVVRYYCTERDKQFRYIALELCSATLQDYVEKKLNFECKIDSVEILRQATMGLSHLHSMDIVHRDVKPHNVLLSMPSGTGEVRAMISDFGLSKKLNIGRVSFSRRSGVTGTDGWIAPEMINGERTTTSIDIFSLGCVFYYVLSRGQHPFGDVLRRQANILTGDYNLDHLDKILPEEELVVVKILIRAMISAKPSARPPCETILKYPMFWCKQSILNFLQDVSDQVESCACGFGAEHTLERGGARVLRGDWRARVCRVVAGDLRTRRTYRGDRVAHLLRAIRNKVPTPTRPHSGLLLPAVLKSIYWVLGYRFRK
Summary
Uniprot
H9ISF3
A0A2H1WVP0
A0A3S2NTW7
A0A212FI45
D6WVT7
A0A1Y1LCF4
+ More
A0A0M9A867 A0A0L7RD49 A0A087ZS85 A0A154PPG3 K7IMS0 A0A232FJS9 A0A2J7QD78 A0A067QMX0 A0A0C9PRE8 A0A2A3E0C8 A0A310SFX9 A0A195CX64 F4WK76 A0A1W4X0U2 A0A1W4XBN9 A0A151JCV0 A0A336M6W9 A0A336M157 A0A151JWF8 A0A158NMN3 N6TY92 U4TV90 E2ABW3 A0A1W4W954 A0A034VU66 Q16QF5 A0A0L0CHF2 A0A0K8UNN4 A0A0A1WHF9 B0W329 B4M4R7 A0A1Q3FEU8 E9I8H9 U5ELH4 B3P081 B3LXS2 A0A1I8PWC0 A0A0M4EG85 A8JR46 A0A0R1E0R2 A0A1V0QBD0 A0A1Q3FBV8 B4JT13 A0A1A9VAE8 A0A3B0KDA0 A0A182GDU1 A0A3B0JNA6 A0A1B0BLV8 W8C6D6 B4K4S4 A0A1A9Y8J0 A0A1B0A230 A0A3B0KIY9 A0A195BWV4 A0A151WHG7 A0A1Q3FEV2 Q293L0 A0A1B0FQP8 B4NEZ0 E0VNS3 A0A1A9WJ68 A0A2M4BAU7 A0A2M4BAR9 A0A2M4BBW0 W5JVI7 A0A2M4A0A7 A0A182FSD4 A0A2M4A0B0 B4GLZ7 A0A2M4A091 A0A2M3YZA8 A0A182NLC8 A0A182JEQ0 A0A2M3ZE88 A0A182P6S0 A0A182QMN2 A0A182M786 A0A182XJ45 A0A1J1IPL6 A0A182UWC3 A0A182JTD1 A0A182L7A8 A0A2M3Z014 A0A340TBI4 A0A182I7R6 B4IHR9 Q5TTF0 A0A1B6MTN9 A0A182RYP4 A0A1B6C1J1 A0A1B6IQ08
A0A0M9A867 A0A0L7RD49 A0A087ZS85 A0A154PPG3 K7IMS0 A0A232FJS9 A0A2J7QD78 A0A067QMX0 A0A0C9PRE8 A0A2A3E0C8 A0A310SFX9 A0A195CX64 F4WK76 A0A1W4X0U2 A0A1W4XBN9 A0A151JCV0 A0A336M6W9 A0A336M157 A0A151JWF8 A0A158NMN3 N6TY92 U4TV90 E2ABW3 A0A1W4W954 A0A034VU66 Q16QF5 A0A0L0CHF2 A0A0K8UNN4 A0A0A1WHF9 B0W329 B4M4R7 A0A1Q3FEU8 E9I8H9 U5ELH4 B3P081 B3LXS2 A0A1I8PWC0 A0A0M4EG85 A8JR46 A0A0R1E0R2 A0A1V0QBD0 A0A1Q3FBV8 B4JT13 A0A1A9VAE8 A0A3B0KDA0 A0A182GDU1 A0A3B0JNA6 A0A1B0BLV8 W8C6D6 B4K4S4 A0A1A9Y8J0 A0A1B0A230 A0A3B0KIY9 A0A195BWV4 A0A151WHG7 A0A1Q3FEV2 Q293L0 A0A1B0FQP8 B4NEZ0 E0VNS3 A0A1A9WJ68 A0A2M4BAU7 A0A2M4BAR9 A0A2M4BBW0 W5JVI7 A0A2M4A0A7 A0A182FSD4 A0A2M4A0B0 B4GLZ7 A0A2M4A091 A0A2M3YZA8 A0A182NLC8 A0A182JEQ0 A0A2M3ZE88 A0A182P6S0 A0A182QMN2 A0A182M786 A0A182XJ45 A0A1J1IPL6 A0A182UWC3 A0A182JTD1 A0A182L7A8 A0A2M3Z014 A0A340TBI4 A0A182I7R6 B4IHR9 Q5TTF0 A0A1B6MTN9 A0A182RYP4 A0A1B6C1J1 A0A1B6IQ08
Pubmed
19121390
22118469
18362917
19820115
28004739
20075255
+ More
28648823 24845553 21719571 21347285 23537049 20798317 25348373 17510324 26108605 25830018 17994087 21282665 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 28356351 26483478 24495485 15632085 20566863 20920257 23761445 20966253 12364791 14747013 17210077
28648823 24845553 21719571 21347285 23537049 20798317 25348373 17510324 26108605 25830018 17994087 21282665 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 28356351 26483478 24495485 15632085 20566863 20920257 23761445 20966253 12364791 14747013 17210077
EMBL
BABH01040313
BABH01040314
BABH01040315
BABH01040316
BABH01040317
ODYU01011405
+ More
SOQ57130.1 RSAL01000200 RVE44453.1 AGBW02008437 OWR53397.1 KQ971358 EFA08613.1 GEZM01062149 GEZM01062148 GEZM01062147 JAV70010.1 KQ435714 KOX79225.1 KQ414615 KOC68783.1 KQ435010 KZC13752.1 NNAY01000124 OXU30749.1 NEVH01015826 PNF26541.1 KK853134 KDR10833.1 GBYB01003858 JAG73625.1 KZ288486 PBC25233.1 KQ760382 OAD60687.1 KQ977231 KYN04749.1 GL888194 EGI65399.1 KQ979038 KYN23486.1 UFQT01000645 SSX26054.1 UFQS01000410 UFQT01000410 SSX03691.1 SSX24056.1 KQ981653 KYN38619.1 ADTU01020633 ADTU01020634 ADTU01020635 ADTU01020636 ADTU01020637 APGK01057050 APGK01057051 KB741277 ENN71262.1 KB631617 ERL84717.1 GL438386 EFN69058.1 GAKP01012953 JAC45999.1 CH477750 EAT36621.1 JRES01000405 KNC31642.1 GDHF01024344 JAI27970.1 GBXI01016499 GBXI01007192 JAC97792.1 JAD07100.1 DS231830 EDS30737.1 CH940652 EDW59628.2 GFDL01008976 JAV26069.1 GL761553 EFZ23129.1 GANO01001406 JAB58465.1 CH954181 EDV48455.1 CH902617 EDV41729.1 CP012526 ALC47341.1 AE014297 ABW08704.1 CM000160 KRK02892.1 KX683314 ARE59261.1 GFDL01010067 JAV24978.1 CH916373 EDV94903.1 OUUW01000008 SPP83706.1 JXUM01056628 JXUM01056629 JXUM01056630 KQ561918 KXJ77167.1 SPP83707.1 JXJN01016586 JXJN01016587 GAMC01008601 JAB97954.1 CH933806 EDW16077.2 SPP83708.1 KQ976395 KYM93114.1 KQ983118 KYQ47270.1 GFDL01008972 JAV26073.1 CM000070 EAL29205.3 CCAG010014772 CH964251 EDW83365.2 DS235346 EEB15029.1 GGFJ01000991 MBW50132.1 GGFJ01000992 MBW50133.1 GGFJ01001372 MBW50513.1 ADMH02000361 ETN66834.1 GGFK01000916 MBW34237.1 GGFK01000902 MBW34223.1 CH479185 EDW38571.1 GGFK01000915 MBW34236.1 GGFM01000851 MBW21602.1 GGFM01006068 MBW26819.1 AXCN02000121 AXCM01003760 CVRI01000056 CRL01658.1 GGFM01001109 MBW21860.1 APCN01002549 CH480840 EDW49436.1 AAAB01008880 EAL40662.4 GEBQ01000680 JAT39297.1 GEDC01030193 GEDC01027232 JAS07105.1 JAS10066.1 GECU01018701 JAS89005.1
SOQ57130.1 RSAL01000200 RVE44453.1 AGBW02008437 OWR53397.1 KQ971358 EFA08613.1 GEZM01062149 GEZM01062148 GEZM01062147 JAV70010.1 KQ435714 KOX79225.1 KQ414615 KOC68783.1 KQ435010 KZC13752.1 NNAY01000124 OXU30749.1 NEVH01015826 PNF26541.1 KK853134 KDR10833.1 GBYB01003858 JAG73625.1 KZ288486 PBC25233.1 KQ760382 OAD60687.1 KQ977231 KYN04749.1 GL888194 EGI65399.1 KQ979038 KYN23486.1 UFQT01000645 SSX26054.1 UFQS01000410 UFQT01000410 SSX03691.1 SSX24056.1 KQ981653 KYN38619.1 ADTU01020633 ADTU01020634 ADTU01020635 ADTU01020636 ADTU01020637 APGK01057050 APGK01057051 KB741277 ENN71262.1 KB631617 ERL84717.1 GL438386 EFN69058.1 GAKP01012953 JAC45999.1 CH477750 EAT36621.1 JRES01000405 KNC31642.1 GDHF01024344 JAI27970.1 GBXI01016499 GBXI01007192 JAC97792.1 JAD07100.1 DS231830 EDS30737.1 CH940652 EDW59628.2 GFDL01008976 JAV26069.1 GL761553 EFZ23129.1 GANO01001406 JAB58465.1 CH954181 EDV48455.1 CH902617 EDV41729.1 CP012526 ALC47341.1 AE014297 ABW08704.1 CM000160 KRK02892.1 KX683314 ARE59261.1 GFDL01010067 JAV24978.1 CH916373 EDV94903.1 OUUW01000008 SPP83706.1 JXUM01056628 JXUM01056629 JXUM01056630 KQ561918 KXJ77167.1 SPP83707.1 JXJN01016586 JXJN01016587 GAMC01008601 JAB97954.1 CH933806 EDW16077.2 SPP83708.1 KQ976395 KYM93114.1 KQ983118 KYQ47270.1 GFDL01008972 JAV26073.1 CM000070 EAL29205.3 CCAG010014772 CH964251 EDW83365.2 DS235346 EEB15029.1 GGFJ01000991 MBW50132.1 GGFJ01000992 MBW50133.1 GGFJ01001372 MBW50513.1 ADMH02000361 ETN66834.1 GGFK01000916 MBW34237.1 GGFK01000902 MBW34223.1 CH479185 EDW38571.1 GGFK01000915 MBW34236.1 GGFM01000851 MBW21602.1 GGFM01006068 MBW26819.1 AXCN02000121 AXCM01003760 CVRI01000056 CRL01658.1 GGFM01001109 MBW21860.1 APCN01002549 CH480840 EDW49436.1 AAAB01008880 EAL40662.4 GEBQ01000680 JAT39297.1 GEDC01030193 GEDC01027232 JAS07105.1 JAS10066.1 GECU01018701 JAS89005.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000007266
UP000053105
UP000053825
+ More
UP000005203 UP000076502 UP000002358 UP000215335 UP000235965 UP000027135 UP000242457 UP000078542 UP000007755 UP000192223 UP000078492 UP000078541 UP000005205 UP000019118 UP000030742 UP000000311 UP000192221 UP000008820 UP000037069 UP000002320 UP000008792 UP000008711 UP000007801 UP000095300 UP000092553 UP000000803 UP000002282 UP000001070 UP000078200 UP000268350 UP000069940 UP000249989 UP000092460 UP000009192 UP000092443 UP000092445 UP000078540 UP000075809 UP000001819 UP000092444 UP000007798 UP000009046 UP000091820 UP000000673 UP000069272 UP000008744 UP000075884 UP000075880 UP000075885 UP000075886 UP000075883 UP000076407 UP000183832 UP000075903 UP000075881 UP000075882 UP000075920 UP000075840 UP000001292 UP000007062 UP000075900
UP000005203 UP000076502 UP000002358 UP000215335 UP000235965 UP000027135 UP000242457 UP000078542 UP000007755 UP000192223 UP000078492 UP000078541 UP000005205 UP000019118 UP000030742 UP000000311 UP000192221 UP000008820 UP000037069 UP000002320 UP000008792 UP000008711 UP000007801 UP000095300 UP000092553 UP000000803 UP000002282 UP000001070 UP000078200 UP000268350 UP000069940 UP000249989 UP000092460 UP000009192 UP000092443 UP000092445 UP000078540 UP000075809 UP000001819 UP000092444 UP000007798 UP000009046 UP000091820 UP000000673 UP000069272 UP000008744 UP000075884 UP000075880 UP000075885 UP000075886 UP000075883 UP000076407 UP000183832 UP000075903 UP000075881 UP000075882 UP000075920 UP000075840 UP000001292 UP000007062 UP000075900
Pfam
Interpro
IPR000719
Prot_kinase_dom
+ More
IPR038357 KEN_sf
IPR018391 PQQ_beta_propeller_repeat
IPR011009 Kinase-like_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR018997 PUB_domain
IPR010513 KEN_dom
IPR008271 Ser/Thr_kinase_AS
IPR033523 IRE2
IPR023801 His_deacetylse_dom
IPR001607 Znf_UBP
IPR000286 His_deacetylse
IPR023696 Ureohydrolase_dom_sf
IPR037138 His_deacetylse_dom_sf
IPR013083 Znf_RING/FYVE/PHD
IPR002372 PQQ_repeat
IPR011044 Quino_amine_DH_bsu
IPR001754 OMPdeCOase_dom
IPR029057 PRTase-like
IPR000836 PRibTrfase_dom
IPR018089 OMPdecase_AS
IPR023031 OPRT
IPR014732 OMPdecase
IPR011060 RibuloseP-bd_barrel
IPR013785 Aldolase_TIM
IPR004467 Or_phspho_trans_dom
IPR038357 KEN_sf
IPR018391 PQQ_beta_propeller_repeat
IPR011009 Kinase-like_dom_sf
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR018997 PUB_domain
IPR010513 KEN_dom
IPR008271 Ser/Thr_kinase_AS
IPR033523 IRE2
IPR023801 His_deacetylse_dom
IPR001607 Znf_UBP
IPR000286 His_deacetylse
IPR023696 Ureohydrolase_dom_sf
IPR037138 His_deacetylse_dom_sf
IPR013083 Znf_RING/FYVE/PHD
IPR002372 PQQ_repeat
IPR011044 Quino_amine_DH_bsu
IPR001754 OMPdeCOase_dom
IPR029057 PRTase-like
IPR000836 PRibTrfase_dom
IPR018089 OMPdecase_AS
IPR023031 OPRT
IPR014732 OMPdecase
IPR011060 RibuloseP-bd_barrel
IPR013785 Aldolase_TIM
IPR004467 Or_phspho_trans_dom
SUPFAM
ProteinModelPortal
H9ISF3
A0A2H1WVP0
A0A3S2NTW7
A0A212FI45
D6WVT7
A0A1Y1LCF4
+ More
A0A0M9A867 A0A0L7RD49 A0A087ZS85 A0A154PPG3 K7IMS0 A0A232FJS9 A0A2J7QD78 A0A067QMX0 A0A0C9PRE8 A0A2A3E0C8 A0A310SFX9 A0A195CX64 F4WK76 A0A1W4X0U2 A0A1W4XBN9 A0A151JCV0 A0A336M6W9 A0A336M157 A0A151JWF8 A0A158NMN3 N6TY92 U4TV90 E2ABW3 A0A1W4W954 A0A034VU66 Q16QF5 A0A0L0CHF2 A0A0K8UNN4 A0A0A1WHF9 B0W329 B4M4R7 A0A1Q3FEU8 E9I8H9 U5ELH4 B3P081 B3LXS2 A0A1I8PWC0 A0A0M4EG85 A8JR46 A0A0R1E0R2 A0A1V0QBD0 A0A1Q3FBV8 B4JT13 A0A1A9VAE8 A0A3B0KDA0 A0A182GDU1 A0A3B0JNA6 A0A1B0BLV8 W8C6D6 B4K4S4 A0A1A9Y8J0 A0A1B0A230 A0A3B0KIY9 A0A195BWV4 A0A151WHG7 A0A1Q3FEV2 Q293L0 A0A1B0FQP8 B4NEZ0 E0VNS3 A0A1A9WJ68 A0A2M4BAU7 A0A2M4BAR9 A0A2M4BBW0 W5JVI7 A0A2M4A0A7 A0A182FSD4 A0A2M4A0B0 B4GLZ7 A0A2M4A091 A0A2M3YZA8 A0A182NLC8 A0A182JEQ0 A0A2M3ZE88 A0A182P6S0 A0A182QMN2 A0A182M786 A0A182XJ45 A0A1J1IPL6 A0A182UWC3 A0A182JTD1 A0A182L7A8 A0A2M3Z014 A0A340TBI4 A0A182I7R6 B4IHR9 Q5TTF0 A0A1B6MTN9 A0A182RYP4 A0A1B6C1J1 A0A1B6IQ08
A0A0M9A867 A0A0L7RD49 A0A087ZS85 A0A154PPG3 K7IMS0 A0A232FJS9 A0A2J7QD78 A0A067QMX0 A0A0C9PRE8 A0A2A3E0C8 A0A310SFX9 A0A195CX64 F4WK76 A0A1W4X0U2 A0A1W4XBN9 A0A151JCV0 A0A336M6W9 A0A336M157 A0A151JWF8 A0A158NMN3 N6TY92 U4TV90 E2ABW3 A0A1W4W954 A0A034VU66 Q16QF5 A0A0L0CHF2 A0A0K8UNN4 A0A0A1WHF9 B0W329 B4M4R7 A0A1Q3FEU8 E9I8H9 U5ELH4 B3P081 B3LXS2 A0A1I8PWC0 A0A0M4EG85 A8JR46 A0A0R1E0R2 A0A1V0QBD0 A0A1Q3FBV8 B4JT13 A0A1A9VAE8 A0A3B0KDA0 A0A182GDU1 A0A3B0JNA6 A0A1B0BLV8 W8C6D6 B4K4S4 A0A1A9Y8J0 A0A1B0A230 A0A3B0KIY9 A0A195BWV4 A0A151WHG7 A0A1Q3FEV2 Q293L0 A0A1B0FQP8 B4NEZ0 E0VNS3 A0A1A9WJ68 A0A2M4BAU7 A0A2M4BAR9 A0A2M4BBW0 W5JVI7 A0A2M4A0A7 A0A182FSD4 A0A2M4A0B0 B4GLZ7 A0A2M4A091 A0A2M3YZA8 A0A182NLC8 A0A182JEQ0 A0A2M3ZE88 A0A182P6S0 A0A182QMN2 A0A182M786 A0A182XJ45 A0A1J1IPL6 A0A182UWC3 A0A182JTD1 A0A182L7A8 A0A2M3Z014 A0A340TBI4 A0A182I7R6 B4IHR9 Q5TTF0 A0A1B6MTN9 A0A182RYP4 A0A1B6C1J1 A0A1B6IQ08
PDB
6HV0
E-value=1.62412e-112,
Score=1042
Ontologies
PATHWAY
GO
GO:0004540
GO:0006397
GO:0016021
GO:0004672
GO:0005524
GO:0005789
GO:0030968
GO:0070059
GO:1990604
GO:0004521
GO:0036498
GO:0004674
GO:0051082
GO:0008270
GO:0006402
GO:0043484
GO:0007030
GO:0061541
GO:0006468
GO:0001751
GO:0034976
GO:0009116
GO:0004590
GO:0044205
GO:0004588
GO:0006207
GO:0004812
GO:0006418
GO:0003723
GO:0046872
GO:0003824
GO:0005515
PANTHER
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.996013
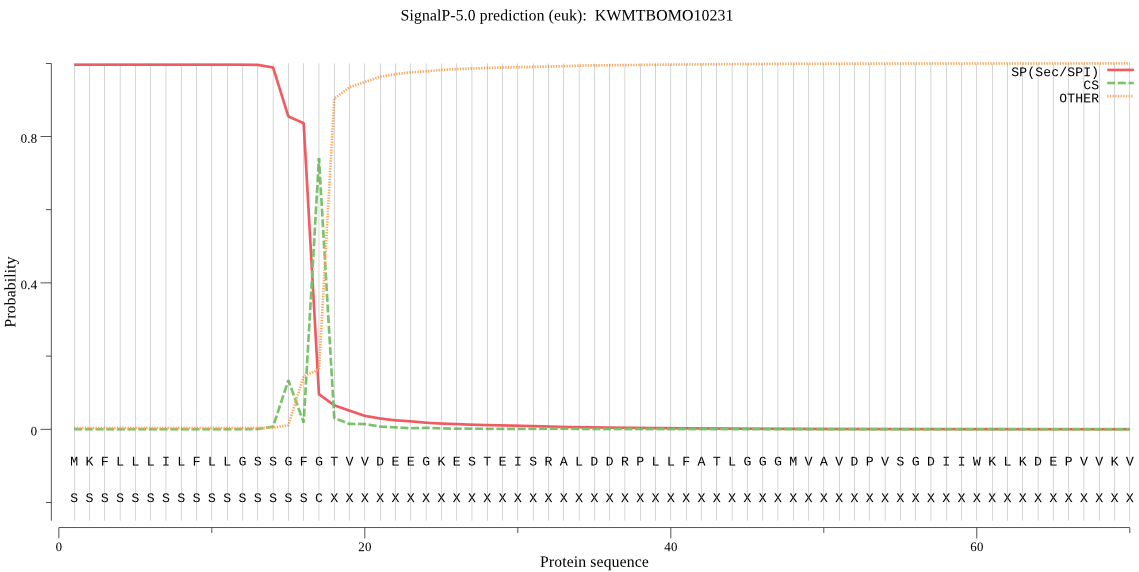
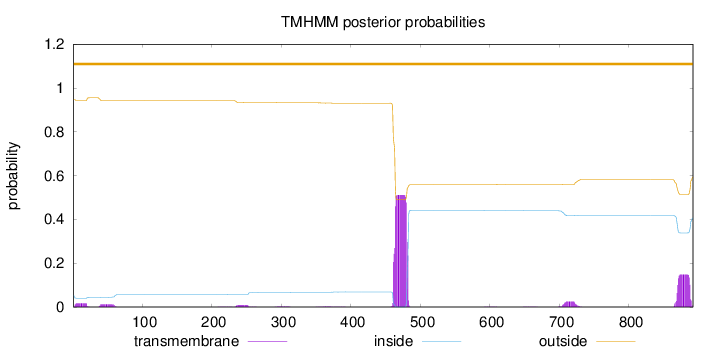
Length:
892
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
14.07498
Exp number, first 60 AAs:
0.57131
Total prob of N-in:
0.05248
outside
1 - 892
Population Genetic Test Statistics
Pi
178.357639
Theta
155.440467
Tajima's D
-1.95567
CLR
0.753879
CSRT
0.0168491575421229
Interpretation
Possibly Positive selection
