Gene
KWMTBOMO10229
Pre Gene Modal
BGIBMGA000176
Annotation
PREDICTED:_testis-expressed_sequence_2_protein-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.645
Sequence
CDS
ATGGAAACGGGCGCAGACGGCACGGCAGAAGGAGAATGGAGCGTCCACGTGAAACATTCTAAAGACAAGCATTCGCATTTGTATCTATTCGCTCGGACTGGACGCGACAAACACGAGTGGTTCCGTAGATTGAGTACGGCAATAGCTGAAGCTAACGCGGCTTCCTCGGACTCGTCCGGTCCCGACGACAAATCGAACGACACGAATGAAAACAAAGACGGCATCGAGCTCGCCGTCTACAAGTTCTCTGAAAAGGACACGGTCGCCTTCGCCAAAACATCGAACAAGTCTGACACCGGCTCCGAGGGTGTTACGATTTCGTCGCTGGGACAAACCCTACCGTCAGATTCGGAAGCCTACGAGAAAAACTTCTGGCCGTATCTGATCAAGATTATACAGCCAAAAAGTCCGTCGCCGGACTGCGAGTGCCGCACCCTGCCCGCGGAACTGACCTGGGTCAACACGGCGCTGGCTCGCGTCGCGTACGACGTGATGCGGGACCCCGTCATCATAGCGCGCGTGCAGAACAGGATACAGAGAAAGCTCAACACTCTCAAGCTGCCGTCGTTCATGTCCCCGCTGACGGTGACGTCACTGTCGCTGGCGGGCGCGTGCCCGGCGCTGGGCGGCGTGGCGGCGCGCTGGGACGCGCGCGGCGTGTGGCTGGACGCCGACCTGCGCTACGACGGCGGCGCCCACATCGCGCTACTCACGCAGATCAACCTCATGAAGCTGAAGGAGAAGAATATGACATTGGAGGAGCAACTCCTCGAGCACAGCGAGAATGTCGTTGAAAGCGACACTTGCGCCACACCGACTTGTGGAGCGTTTTCTGAAAAACAACTCAGAAAAAGGAAACCCGCGATATACGATTCCGATATAGAAGACTCCGCGGAGTCGAGCAGCGACGACGAGAGTCCGCCGGTGCAGCCCGTCGACAGCGCTGACAACCTAACGGTATCGGAGTCTCCATCTACGACGACGGAAGGTGGATCGTCCAAAAAGAAATTCTTACGGATGGTCGATAAGATTGCAACCAACAAATACTTCCAGCAGGTAACCGACTACAAGTACGTGAAGCGTGCAATGGAGGGACTGTCCAACACGGATATAAAACTGCATCTAGAAATACAAGGTCTGGAGGGACGCCTGTCATTTAATCTCCCGCCTCCGCCGCACGATCGAGTATGGATCGGTTTCCGAACGAATCCTCAACTGGTGCTAAAAGCGCGTCCCGCGGTCGGCGCGCGCACATTACGTTTCACGCACATCTCCAACTGGATCGAGCAGAAGCTGAGCAAAGAATTCGAAAAGGTCCTCGTTTTGCCGAACATGGAGGATATCATTATCGATATAATGACGCCAACACCGGTGCAGTTCGAGTGA
Protein
METGADGTAEGEWSVHVKHSKDKHSHLYLFARTGRDKHEWFRRLSTAIAEANAASSDSSGPDDKSNDTNENKDGIELAVYKFSEKDTVAFAKTSNKSDTGSEGVTISSLGQTLPSDSEAYEKNFWPYLIKIIQPKSPSPDCECRTLPAELTWVNTALARVAYDVMRDPVIIARVQNRIQRKLNTLKLPSFMSPLTVTSLSLAGACPALGGVAARWDARGVWLDADLRYDGGAHIALLTQINLMKLKEKNMTLEEQLLEHSENVVESDTCATPTCGAFSEKQLRKRKPAIYDSDIEDSAESSSDDESPPVQPVDSADNLTVSESPSTTTEGGSSKKKFLRMVDKIATNKYFQQVTDYKYVKRAMEGLSNTDIKLHLEIQGLEGRLSFNLPPPPHDRVWIGFRTNPQLVLKARPAVGARTLRFTHISNWIEQKLSKEFEKVLVLPNMEDIIIDIMTPTPVQFE
Summary
Uniprot
A0A194QB78
A0A2H1WVU9
A0A212FI41
A0A2A4J0N5
A0A1E1W1H1
A0A1E1WL35
+ More
A0A1E1VZC9 N6SS96 V5I8V4 A0A232EDN6 A0A067QXG5 K7IWQ1 A0A026WAH6 A0A1Y1M6M5 A0A2C9K296 R7U2I9 A0A0N1IU13 A0A151N286 A0A3Q3BNT8 A0A0J7L6N2 A0A151J6J0 A0A195FLC4 F4WJ97 A0A3L8RXL0 A0A087UJD1 A0A091K7X7 A0A0L7RCR5 A0A195CCS1 A0A3Q3WAD6 A0A151WSL0 A0A0S7JNB6 A0A1U7S478 U3JL05 A0A3B4TDE0 A0A091G7G5 A0A091ELX6 A0A3Q3KDX7 A0A3P9BYJ4 A0A195BVD0 A0A158NEC0 A0A3Q4AW03 A0A3B4WNE6 A0A3M0KXV1 H0Z8M1 A0A3B4YKF7 A0A3Q2U9T7 E2BLQ7 A0A2A3E129 A0A146RG23 A0A3B5M5W4 A0A0S7JNA8 A0A091VA32 A0A146ZJK0 A0A3Q2WQG5 A0A094KMC5 A0A3Q2WSP9 A0A3Q2VS76 I3J635 A0A3Q2VJB1 A0A218UA70 A0A1V4KSW8 A0A091J8N3 A0A3P8PNB9 A0A3P9BZB3 A0A1V4KSS9 K1RZX4 A0A3P9MAV9 F1NX58 A0A3B4U3G9 A0A2I0LTF3 A0A0S7JPI0 F1QF95 R7VPC0 A0A3P9HZ95 A5D6U2 A0A3P9KYD0 E2AEI5 A0A3Q3LI50 F6WP91 A0A147B2J8 A0A3P9HZ42 T1KB68 A0A093P1F5 A0A3B3HBR1 A0A087RG61 A0A3B3RS43 A0A146MMV4 H2L955 A0A3Q4HMP8 A0A3Q4HR51 A0A3B3H9H1 A0A091MDH4
A0A1E1VZC9 N6SS96 V5I8V4 A0A232EDN6 A0A067QXG5 K7IWQ1 A0A026WAH6 A0A1Y1M6M5 A0A2C9K296 R7U2I9 A0A0N1IU13 A0A151N286 A0A3Q3BNT8 A0A0J7L6N2 A0A151J6J0 A0A195FLC4 F4WJ97 A0A3L8RXL0 A0A087UJD1 A0A091K7X7 A0A0L7RCR5 A0A195CCS1 A0A3Q3WAD6 A0A151WSL0 A0A0S7JNB6 A0A1U7S478 U3JL05 A0A3B4TDE0 A0A091G7G5 A0A091ELX6 A0A3Q3KDX7 A0A3P9BYJ4 A0A195BVD0 A0A158NEC0 A0A3Q4AW03 A0A3B4WNE6 A0A3M0KXV1 H0Z8M1 A0A3B4YKF7 A0A3Q2U9T7 E2BLQ7 A0A2A3E129 A0A146RG23 A0A3B5M5W4 A0A0S7JNA8 A0A091VA32 A0A146ZJK0 A0A3Q2WQG5 A0A094KMC5 A0A3Q2WSP9 A0A3Q2VS76 I3J635 A0A3Q2VJB1 A0A218UA70 A0A1V4KSW8 A0A091J8N3 A0A3P8PNB9 A0A3P9BZB3 A0A1V4KSS9 K1RZX4 A0A3P9MAV9 F1NX58 A0A3B4U3G9 A0A2I0LTF3 A0A0S7JPI0 F1QF95 R7VPC0 A0A3P9HZ95 A5D6U2 A0A3P9KYD0 E2AEI5 A0A3Q3LI50 F6WP91 A0A147B2J8 A0A3P9HZ42 T1KB68 A0A093P1F5 A0A3B3HBR1 A0A087RG61 A0A3B3RS43 A0A146MMV4 H2L955 A0A3Q4HMP8 A0A3Q4HR51 A0A3B3H9H1 A0A091MDH4
Pubmed
EMBL
KQ459232
KPJ02717.1
ODYU01011385
SOQ57096.1
AGBW02008437
OWR53398.1
+ More
NWSH01003881 PCG65727.1 GDQN01010204 JAT80850.1 GDQN01003406 JAT87648.1 GDQN01010967 JAT80087.1 APGK01058603 KB741291 ENN70519.1 GALX01004030 JAB64436.1 NNAY01006053 OXU16476.1 KK853134 KDR10832.1 AAZX01007094 KK107356 QOIP01000010 EZA52034.1 RLU17469.1 GEZM01040995 JAV80668.1 AMQN01010923 KB308570 ELT97380.1 KQ435714 KOX79224.1 AKHW03004153 KYO30837.1 LBMM01000514 KMQ98164.1 KQ979851 KYN18834.1 KQ981490 KYN41062.1 GL888182 EGI65675.1 QUSF01000142 RLV89616.1 KK120076 KFM77470.1 KK546375 KFP32461.1 KQ414615 KOC68782.1 KQ978023 KYM97883.1 KQ982769 KYQ50818.1 GBYX01234347 JAO66657.1 AGTO01000599 KL447789 KFO77114.1 KK718885 KFO58948.1 KQ976401 KYM92537.1 ADTU01013242 QRBI01000098 RMC17531.1 ABQF01123824 AADN05000376 GL449036 EFN83391.1 KZ288486 PBC25234.1 GCES01119266 JAQ67056.1 GBYX01234344 JAO66660.1 KL410807 KFQ99933.1 GCES01020301 JAR66022.1 KL337247 KFZ50705.1 AERX01019107 AERX01019108 MUZQ01000541 OWK50481.1 LSYS01001700 OPJ87456.1 KK501727 KFP17359.1 OPJ87455.1 JH818947 EKC40551.1 AKCR02000102 PKK20715.1 GBYX01234346 JAO66658.1 CR392029 LO018123 KB375873 EMC80193.1 BC139885 AAI39886.1 GL438862 EFN68142.1 AAMC01050966 AAMC01050967 AAMC01050968 GCES01001721 JAR84602.1 CAEY01001945 KL225320 KFW70868.1 KL226347 KFM12465.1 GCES01165950 JAQ20372.1 KK527948 KFP68812.1
NWSH01003881 PCG65727.1 GDQN01010204 JAT80850.1 GDQN01003406 JAT87648.1 GDQN01010967 JAT80087.1 APGK01058603 KB741291 ENN70519.1 GALX01004030 JAB64436.1 NNAY01006053 OXU16476.1 KK853134 KDR10832.1 AAZX01007094 KK107356 QOIP01000010 EZA52034.1 RLU17469.1 GEZM01040995 JAV80668.1 AMQN01010923 KB308570 ELT97380.1 KQ435714 KOX79224.1 AKHW03004153 KYO30837.1 LBMM01000514 KMQ98164.1 KQ979851 KYN18834.1 KQ981490 KYN41062.1 GL888182 EGI65675.1 QUSF01000142 RLV89616.1 KK120076 KFM77470.1 KK546375 KFP32461.1 KQ414615 KOC68782.1 KQ978023 KYM97883.1 KQ982769 KYQ50818.1 GBYX01234347 JAO66657.1 AGTO01000599 KL447789 KFO77114.1 KK718885 KFO58948.1 KQ976401 KYM92537.1 ADTU01013242 QRBI01000098 RMC17531.1 ABQF01123824 AADN05000376 GL449036 EFN83391.1 KZ288486 PBC25234.1 GCES01119266 JAQ67056.1 GBYX01234344 JAO66660.1 KL410807 KFQ99933.1 GCES01020301 JAR66022.1 KL337247 KFZ50705.1 AERX01019107 AERX01019108 MUZQ01000541 OWK50481.1 LSYS01001700 OPJ87456.1 KK501727 KFP17359.1 OPJ87455.1 JH818947 EKC40551.1 AKCR02000102 PKK20715.1 GBYX01234346 JAO66658.1 CR392029 LO018123 KB375873 EMC80193.1 BC139885 AAI39886.1 GL438862 EFN68142.1 AAMC01050966 AAMC01050967 AAMC01050968 GCES01001721 JAR84602.1 CAEY01001945 KL225320 KFW70868.1 KL226347 KFM12465.1 GCES01165950 JAQ20372.1 KK527948 KFP68812.1
Proteomes
UP000053268
UP000007151
UP000218220
UP000019118
UP000215335
UP000027135
+ More
UP000002358 UP000053097 UP000279307 UP000076420 UP000014760 UP000053105 UP000050525 UP000264800 UP000036403 UP000078492 UP000078541 UP000007755 UP000276834 UP000054359 UP000053825 UP000078542 UP000261620 UP000075809 UP000189705 UP000016665 UP000261420 UP000053760 UP000052976 UP000261600 UP000265160 UP000078540 UP000005205 UP000261360 UP000269221 UP000007754 UP000000539 UP000008237 UP000242457 UP000261380 UP000053283 UP000264840 UP000005207 UP000197619 UP000190648 UP000053119 UP000265100 UP000005408 UP000265180 UP000053872 UP000000437 UP000265200 UP000000311 UP000261640 UP000008143 UP000015104 UP000054081 UP000001038 UP000053286 UP000261540 UP000261580
UP000002358 UP000053097 UP000279307 UP000076420 UP000014760 UP000053105 UP000050525 UP000264800 UP000036403 UP000078492 UP000078541 UP000007755 UP000276834 UP000054359 UP000053825 UP000078542 UP000261620 UP000075809 UP000189705 UP000016665 UP000261420 UP000053760 UP000052976 UP000261600 UP000265160 UP000078540 UP000005205 UP000261360 UP000269221 UP000007754 UP000000539 UP000008237 UP000242457 UP000261380 UP000053283 UP000264840 UP000005207 UP000197619 UP000190648 UP000053119 UP000265100 UP000005408 UP000265180 UP000053872 UP000000437 UP000265200 UP000000311 UP000261640 UP000008143 UP000015104 UP000054081 UP000001038 UP000053286 UP000261540 UP000261580
PRIDE
Interpro
ProteinModelPortal
A0A194QB78
A0A2H1WVU9
A0A212FI41
A0A2A4J0N5
A0A1E1W1H1
A0A1E1WL35
+ More
A0A1E1VZC9 N6SS96 V5I8V4 A0A232EDN6 A0A067QXG5 K7IWQ1 A0A026WAH6 A0A1Y1M6M5 A0A2C9K296 R7U2I9 A0A0N1IU13 A0A151N286 A0A3Q3BNT8 A0A0J7L6N2 A0A151J6J0 A0A195FLC4 F4WJ97 A0A3L8RXL0 A0A087UJD1 A0A091K7X7 A0A0L7RCR5 A0A195CCS1 A0A3Q3WAD6 A0A151WSL0 A0A0S7JNB6 A0A1U7S478 U3JL05 A0A3B4TDE0 A0A091G7G5 A0A091ELX6 A0A3Q3KDX7 A0A3P9BYJ4 A0A195BVD0 A0A158NEC0 A0A3Q4AW03 A0A3B4WNE6 A0A3M0KXV1 H0Z8M1 A0A3B4YKF7 A0A3Q2U9T7 E2BLQ7 A0A2A3E129 A0A146RG23 A0A3B5M5W4 A0A0S7JNA8 A0A091VA32 A0A146ZJK0 A0A3Q2WQG5 A0A094KMC5 A0A3Q2WSP9 A0A3Q2VS76 I3J635 A0A3Q2VJB1 A0A218UA70 A0A1V4KSW8 A0A091J8N3 A0A3P8PNB9 A0A3P9BZB3 A0A1V4KSS9 K1RZX4 A0A3P9MAV9 F1NX58 A0A3B4U3G9 A0A2I0LTF3 A0A0S7JPI0 F1QF95 R7VPC0 A0A3P9HZ95 A5D6U2 A0A3P9KYD0 E2AEI5 A0A3Q3LI50 F6WP91 A0A147B2J8 A0A3P9HZ42 T1KB68 A0A093P1F5 A0A3B3HBR1 A0A087RG61 A0A3B3RS43 A0A146MMV4 H2L955 A0A3Q4HMP8 A0A3Q4HR51 A0A3B3H9H1 A0A091MDH4
A0A1E1VZC9 N6SS96 V5I8V4 A0A232EDN6 A0A067QXG5 K7IWQ1 A0A026WAH6 A0A1Y1M6M5 A0A2C9K296 R7U2I9 A0A0N1IU13 A0A151N286 A0A3Q3BNT8 A0A0J7L6N2 A0A151J6J0 A0A195FLC4 F4WJ97 A0A3L8RXL0 A0A087UJD1 A0A091K7X7 A0A0L7RCR5 A0A195CCS1 A0A3Q3WAD6 A0A151WSL0 A0A0S7JNB6 A0A1U7S478 U3JL05 A0A3B4TDE0 A0A091G7G5 A0A091ELX6 A0A3Q3KDX7 A0A3P9BYJ4 A0A195BVD0 A0A158NEC0 A0A3Q4AW03 A0A3B4WNE6 A0A3M0KXV1 H0Z8M1 A0A3B4YKF7 A0A3Q2U9T7 E2BLQ7 A0A2A3E129 A0A146RG23 A0A3B5M5W4 A0A0S7JNA8 A0A091VA32 A0A146ZJK0 A0A3Q2WQG5 A0A094KMC5 A0A3Q2WSP9 A0A3Q2VS76 I3J635 A0A3Q2VJB1 A0A218UA70 A0A1V4KSW8 A0A091J8N3 A0A3P8PNB9 A0A3P9BZB3 A0A1V4KSS9 K1RZX4 A0A3P9MAV9 F1NX58 A0A3B4U3G9 A0A2I0LTF3 A0A0S7JPI0 F1QF95 R7VPC0 A0A3P9HZ95 A5D6U2 A0A3P9KYD0 E2AEI5 A0A3Q3LI50 F6WP91 A0A147B2J8 A0A3P9HZ42 T1KB68 A0A093P1F5 A0A3B3HBR1 A0A087RG61 A0A3B3RS43 A0A146MMV4 H2L955 A0A3Q4HMP8 A0A3Q4HR51 A0A3B3H9H1 A0A091MDH4
Ontologies
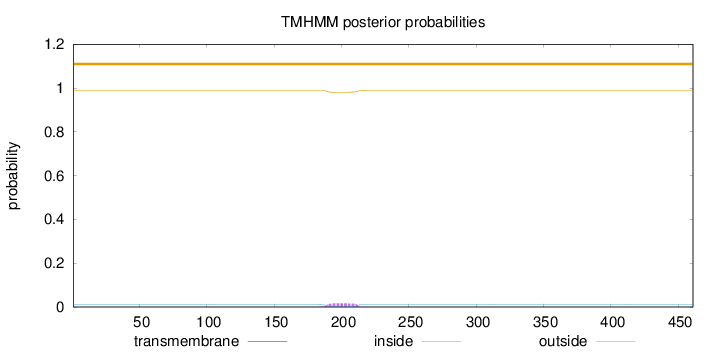
Topology
Length:
461
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.38546
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01207
outside
1 - 461
Population Genetic Test Statistics
Pi
216.668602
Theta
172.837209
Tajima's D
0.755256
CLR
0.748472
CSRT
0.587270636468177
Interpretation
Uncertain
