Gene
KWMTBOMO10225
Pre Gene Modal
BGIBMGA000178
Annotation
PREDICTED:_UNC93-like_protein_MFSD11_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.591
Sequence
CDS
ATGGGAAATATTGAGAAAACTATACTCGACAGTATTACGCAAGACGACAGTTCCTTCACCGGAGATGGGTACACATCGCTCGCCATTATCTACGCTACTCTCGCAATTTGCAACTGGTTGGCACCATCGGCTATTACGATTACGGGTCCAAGAGGGGCAATGCTTATCGGGGCCGTCACATATTTGTTCTTCATAGTGACGTTCCTGTTTCCTCGCACGTGGCTGCTGTACGTGGCGAGCGTGCTGATCGGCGCCGGGGCAGCCATGATATGGACCGGCCAAGGGAACTATCTCACAATGAACAGCGACGCGGAAACGATATCGAGGAACTCGGGTATCTTCTGGGCTATGCTACAATGCAGCATGTTCTTCGGTAATCTGTTCGTGTTCATAAAGTTCCAAGGGAAATCTCATATTGATTTGGAGACGCGCAACGTTGTGTTCGGCGCCCTGACCGGCGTGTGCGTCCTCGGGATCGTCTTCCTCGCGCTACTGCGGCCGATCAAGCGGACGACCACCGTCGAAGACGCCAAGGTTGAAATCGCCTATCGCGACGCCGAAGGGCCGCTGGAGGCTTTCAAGGGCGCCCTGAAATTATTCTGCACCAAAGATATGCTACTTCTGAGCGTCACCTTCATCTATACCGGAATCGAGCTTTCGTTCTTCAGCGGCGTGTACAGTCCCAGTATCGGGTTCACTTTGGCGATGGGGGAAAACGCGAAGCAGCTCGTCGGGCTCTCTGGGGTGTTCATTGGAATGGGCGAAGTTCTCGGCGGGGCTTTGTTCGGTATTTTGGGAAGCAAGACCACACGTTGGGGCCGCGATCCCATCGTCATCATGGGCTTCATAATCCACCTCAGCAGTTTCTTCCTGATCTTCATTAACTTGCCGACGGTGGCGCCCTTCGGGGACACGTCCGAGGTGTCTTACATCACCCCGTCTGCTGAGCTGGCGATGTTCTGCAGCTTCCTGCTGGGCTTCGGAGACGCTTGCTTCAACACTCAGATCTACTCGATACTCGGCGGCAATTACTCGGATAACAGCACGTCCGCGTTTGCGCTCTTCAAGTTTACACAGTCTCTGGCGGCGGCGGCGTGTTTCTTCTACTCGTCGAAGGCGCTGCTGTCGGTGCAGCTCGTGGTGCTCGCCGCGCTGGCGTCCGTCGGCACGGCGTGCTTCTGCCGCGTGGAGTGGGCGGGCAGGGCGCGCGCGCGGGCCGCCGCCCTCGCCGCCGCCGCCGACGACAAACCCTCGCCCTCGCACGCTCTCGCCGACGCCGACGCCGACAACGCTCTGCACTACGACTGA
Protein
MGNIEKTILDSITQDDSSFTGDGYTSLAIIYATLAICNWLAPSAITITGPRGAMLIGAVTYLFFIVTFLFPRTWLLYVASVLIGAGAAMIWTGQGNYLTMNSDAETISRNSGIFWAMLQCSMFFGNLFVFIKFQGKSHIDLETRNVVFGALTGVCVLGIVFLALLRPIKRTTTVEDAKVEIAYRDAEGPLEAFKGALKLFCTKDMLLLSVTFIYTGIELSFFSGVYSPSIGFTLAMGENAKQLVGLSGVFIGMGEVLGGALFGILGSKTTRWGRDPIVIMGFIIHLSSFFLIFINLPTVAPFGDTSEVSYITPSAELAMFCSFLLGFGDACFNTQIYSILGGNYSDNSTSAFALFKFTQSLAAAACFFYSSKALLSVQLVVLAALASVGTACFCRVEWAGRARARAAALAAAADDKPSPSHALADADADNALHYD
Summary
Uniprot
A0A194QT90
A0A194QAZ4
A0A2A4J296
A0A212FFE0
A0A2J7QD95
A0A1Y1N3N7
+ More
W8ATJ8 D6WE38 A0A0T6BCT8 A0A1L8EFU0 A0A336MQ22 A0A1L8EFE2 A0A1S4FYZ9 A0A1L8EFH7 Q16JK3 A0A1L8EFE4 A0A1L8EFL8 U5EXR4 T1PIZ3 A0A1I8N4W1 A0A0L0C0S7 A0A1I8NV62 A0A0K8VHX8 A0A1Q3G301 A0A0K8UME6 A0A0A1XH39 A0A0A1X4A1 A0A0M4EV67 W5JGJ0 A0A1B6JVR4 A0A1B6FCW5 A0A1B6DK71 A0A084VNX5 A0A182IAN3 A0A1W4XD41 A0A2P2I6R7 A0A182XBV1 A0A182UUX2 A0A1S4ECY5 Q7QF54 B4M0B9 B4JRQ6 A0A0P4VRA8 A0A182IKW5 A0A182NC31 B4K746 A0A182MJD4 T1HPI4 T1D6C8 A0A182QIM8 A0A182VV65 A0A182K5P8 A0A182YR78 A0A0A9Z2Z0 A0A0A9YX13 A0A182RM61 E9FSV2 B5DVK2 B4NJQ4 A0A3B0JIF3 A0A182F8E4 A0A224XGX1 A0A1W4UMT2 A0A023F2F4 B4QSA1 B4HGX0 Q9VG64 B3M2R9 B3P4B1 B4PPA7 A0A182LM32 A0A0P5G8L1 A0A0P5S9Y5 A0A0P5RY90 A0A0P5LAZ5 A0A0P5N0F0 A0A0P5PTC8 A0A162D8Z8 A0A0P5K4Y7 A0A0P5LHK0 A0A0P5K3P3 A0A023EQW2 A0A0P5IVW9 A0A0P5P1T4 A0A0P5G7R1 J3JTV8 A0A0N8BRR6 A0A0P6ERD2 A0A0N8BQQ4 N6TWH6 A0A0P4Y8Y4 A0A0P5BCL7 A0A293N3C7 A0A0P5VMA1 A0A0P5NX51 A0A0P5Z9B7 A0A0P4Z7K9 A0A0P5GSV1 A0A0P5ZFL4 U4UBS0
W8ATJ8 D6WE38 A0A0T6BCT8 A0A1L8EFU0 A0A336MQ22 A0A1L8EFE2 A0A1S4FYZ9 A0A1L8EFH7 Q16JK3 A0A1L8EFE4 A0A1L8EFL8 U5EXR4 T1PIZ3 A0A1I8N4W1 A0A0L0C0S7 A0A1I8NV62 A0A0K8VHX8 A0A1Q3G301 A0A0K8UME6 A0A0A1XH39 A0A0A1X4A1 A0A0M4EV67 W5JGJ0 A0A1B6JVR4 A0A1B6FCW5 A0A1B6DK71 A0A084VNX5 A0A182IAN3 A0A1W4XD41 A0A2P2I6R7 A0A182XBV1 A0A182UUX2 A0A1S4ECY5 Q7QF54 B4M0B9 B4JRQ6 A0A0P4VRA8 A0A182IKW5 A0A182NC31 B4K746 A0A182MJD4 T1HPI4 T1D6C8 A0A182QIM8 A0A182VV65 A0A182K5P8 A0A182YR78 A0A0A9Z2Z0 A0A0A9YX13 A0A182RM61 E9FSV2 B5DVK2 B4NJQ4 A0A3B0JIF3 A0A182F8E4 A0A224XGX1 A0A1W4UMT2 A0A023F2F4 B4QSA1 B4HGX0 Q9VG64 B3M2R9 B3P4B1 B4PPA7 A0A182LM32 A0A0P5G8L1 A0A0P5S9Y5 A0A0P5RY90 A0A0P5LAZ5 A0A0P5N0F0 A0A0P5PTC8 A0A162D8Z8 A0A0P5K4Y7 A0A0P5LHK0 A0A0P5K3P3 A0A023EQW2 A0A0P5IVW9 A0A0P5P1T4 A0A0P5G7R1 J3JTV8 A0A0N8BRR6 A0A0P6ERD2 A0A0N8BQQ4 N6TWH6 A0A0P4Y8Y4 A0A0P5BCL7 A0A293N3C7 A0A0P5VMA1 A0A0P5NX51 A0A0P5Z9B7 A0A0P4Z7K9 A0A0P5GSV1 A0A0P5ZFL4 U4UBS0
Pubmed
26354079
22118469
28004739
24495485
18362917
19820115
+ More
17510324 25315136 26108605 25830018 20920257 23761445 24438588 12364791 14747013 17210077 17994087 27129103 24330624 25244985 25401762 26823975 21292972 15632085 25474469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20966253 24945155 22516182 23537049
17510324 25315136 26108605 25830018 20920257 23761445 24438588 12364791 14747013 17210077 17994087 27129103 24330624 25244985 25401762 26823975 21292972 15632085 25474469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20966253 24945155 22516182 23537049
EMBL
KQ461155
KPJ08220.1
KQ459232
KPJ02713.1
NWSH01004089
PCG65503.1
+ More
AGBW02008828 OWR52451.1 NEVH01015826 PNF26549.1 GEZM01013587 JAV92449.1 GAMC01014385 JAB92170.1 KQ971318 EFA00373.1 LJIG01001778 KRT85162.1 GFDG01001356 JAV17443.1 UFQT01002002 SSX32366.1 GFDG01001359 JAV17440.1 GFDG01001358 JAV17441.1 CH478005 EAT34452.1 GFDG01001357 JAV17442.1 GFDG01001355 JAV17444.1 GANO01000815 JAB59056.1 KA648085 AFP62714.1 JRES01001062 KNC25900.1 GDHF01013852 GDHF01006710 JAI38462.1 JAI45604.1 GFDL01000863 JAV34182.1 GDHF01024641 GDHF01010155 JAI27673.1 JAI42159.1 GBXI01004020 JAD10272.1 GBXI01008370 JAD05922.1 CP012526 ALC47142.1 ADMH02001252 ETN63477.1 GECU01024584 GECU01015551 GECU01004399 GECU01003709 JAS83122.1 JAS92155.1 JAT03308.1 JAT03998.1 GECZ01021727 GECZ01021453 JAS48042.1 JAS48316.1 GEDC01011288 JAS26010.1 ATLV01014940 KE524996 KFB39669.1 APCN01001282 IACF01004017 LAB69616.1 AAAB01008846 EAA06364.3 CH940650 EDW67281.1 CH916373 EDV94446.1 GDKW01001656 JAI54939.1 CH933806 EDW16359.1 AXCM01001560 ACPB03010547 ACPB03010548 GALA01000118 JAA94734.1 AXCN02001428 GBHO01007444 GDHC01015939 GDHC01004745 JAG36160.1 JAQ02690.1 JAQ13884.1 GBHO01007443 GBRD01002853 JAG36161.1 JAG62968.1 GL732524 EFX89753.1 CM000070 EDY67669.1 CH964272 EDW85016.1 OUUW01000005 SPP80503.1 GFTR01006152 JAW10274.1 GBBI01003242 JAC15470.1 CM000364 EDX13197.1 CH480815 EDW42441.1 AE014297 AY071334 AAF54824.2 AAL48956.1 CH902617 EDV42390.2 CH954181 EDV49356.1 CM000160 EDW97113.1 KRK03535.1 GDIQ01245094 JAK06631.1 GDIQ01094741 JAL56985.1 GDIQ01094740 JAL56986.1 GDIQ01176911 JAK74814.1 GDIQ01205611 GDIQ01156775 GDIQ01144185 GDIQ01127291 GDIQ01106744 GDIQ01102501 GDIQ01044322 GDIQ01039176 JAK94950.1 GDIQ01123741 JAL27985.1 LRGB01003123 KZS04435.1 GDIQ01194657 JAK57068.1 GDIQ01194658 JAK57067.1 GDIQ01190121 JAK61604.1 GAPW01002083 JAC11515.1 GDIQ01208314 GDIQ01170275 GDIQ01108991 GDIQ01067466 GDIQ01050874 GDIQ01046369 JAK43411.1 GDIQ01136381 JAL15345.1 GDIQ01245093 JAK06632.1 BT126667 AEE61630.1 GDIQ01151331 JAL00395.1 GDIQ01072360 JAN22377.1 GDIQ01154227 JAK97498.1 APGK01049736 APGK01049737 KB741156 ENN73610.1 GDIP01233152 JAI90249.1 GDIP01191693 JAJ31709.1 GFWV01022897 MAA47624.1 GDIP01099356 JAM04359.1 GDIQ01140942 JAL10784.1 GDIP01059702 JAM44013.1 GDIP01216731 JAJ06671.1 GDIQ01236904 JAK14821.1 GDIP01045364 JAM58351.1 KB631941 ERL87385.1
AGBW02008828 OWR52451.1 NEVH01015826 PNF26549.1 GEZM01013587 JAV92449.1 GAMC01014385 JAB92170.1 KQ971318 EFA00373.1 LJIG01001778 KRT85162.1 GFDG01001356 JAV17443.1 UFQT01002002 SSX32366.1 GFDG01001359 JAV17440.1 GFDG01001358 JAV17441.1 CH478005 EAT34452.1 GFDG01001357 JAV17442.1 GFDG01001355 JAV17444.1 GANO01000815 JAB59056.1 KA648085 AFP62714.1 JRES01001062 KNC25900.1 GDHF01013852 GDHF01006710 JAI38462.1 JAI45604.1 GFDL01000863 JAV34182.1 GDHF01024641 GDHF01010155 JAI27673.1 JAI42159.1 GBXI01004020 JAD10272.1 GBXI01008370 JAD05922.1 CP012526 ALC47142.1 ADMH02001252 ETN63477.1 GECU01024584 GECU01015551 GECU01004399 GECU01003709 JAS83122.1 JAS92155.1 JAT03308.1 JAT03998.1 GECZ01021727 GECZ01021453 JAS48042.1 JAS48316.1 GEDC01011288 JAS26010.1 ATLV01014940 KE524996 KFB39669.1 APCN01001282 IACF01004017 LAB69616.1 AAAB01008846 EAA06364.3 CH940650 EDW67281.1 CH916373 EDV94446.1 GDKW01001656 JAI54939.1 CH933806 EDW16359.1 AXCM01001560 ACPB03010547 ACPB03010548 GALA01000118 JAA94734.1 AXCN02001428 GBHO01007444 GDHC01015939 GDHC01004745 JAG36160.1 JAQ02690.1 JAQ13884.1 GBHO01007443 GBRD01002853 JAG36161.1 JAG62968.1 GL732524 EFX89753.1 CM000070 EDY67669.1 CH964272 EDW85016.1 OUUW01000005 SPP80503.1 GFTR01006152 JAW10274.1 GBBI01003242 JAC15470.1 CM000364 EDX13197.1 CH480815 EDW42441.1 AE014297 AY071334 AAF54824.2 AAL48956.1 CH902617 EDV42390.2 CH954181 EDV49356.1 CM000160 EDW97113.1 KRK03535.1 GDIQ01245094 JAK06631.1 GDIQ01094741 JAL56985.1 GDIQ01094740 JAL56986.1 GDIQ01176911 JAK74814.1 GDIQ01205611 GDIQ01156775 GDIQ01144185 GDIQ01127291 GDIQ01106744 GDIQ01102501 GDIQ01044322 GDIQ01039176 JAK94950.1 GDIQ01123741 JAL27985.1 LRGB01003123 KZS04435.1 GDIQ01194657 JAK57068.1 GDIQ01194658 JAK57067.1 GDIQ01190121 JAK61604.1 GAPW01002083 JAC11515.1 GDIQ01208314 GDIQ01170275 GDIQ01108991 GDIQ01067466 GDIQ01050874 GDIQ01046369 JAK43411.1 GDIQ01136381 JAL15345.1 GDIQ01245093 JAK06632.1 BT126667 AEE61630.1 GDIQ01151331 JAL00395.1 GDIQ01072360 JAN22377.1 GDIQ01154227 JAK97498.1 APGK01049736 APGK01049737 KB741156 ENN73610.1 GDIP01233152 JAI90249.1 GDIP01191693 JAJ31709.1 GFWV01022897 MAA47624.1 GDIP01099356 JAM04359.1 GDIQ01140942 JAL10784.1 GDIP01059702 JAM44013.1 GDIP01216731 JAJ06671.1 GDIQ01236904 JAK14821.1 GDIP01045364 JAM58351.1 KB631941 ERL87385.1
Proteomes
UP000053240
UP000053268
UP000218220
UP000007151
UP000235965
UP000007266
+ More
UP000008820 UP000095301 UP000037069 UP000095300 UP000092553 UP000000673 UP000030765 UP000075840 UP000192223 UP000076407 UP000075903 UP000079169 UP000007062 UP000008792 UP000001070 UP000075880 UP000075884 UP000009192 UP000075883 UP000015103 UP000075886 UP000075920 UP000075881 UP000076408 UP000075900 UP000000305 UP000001819 UP000007798 UP000268350 UP000069272 UP000192221 UP000000304 UP000001292 UP000000803 UP000007801 UP000008711 UP000002282 UP000075882 UP000076858 UP000019118 UP000030742
UP000008820 UP000095301 UP000037069 UP000095300 UP000092553 UP000000673 UP000030765 UP000075840 UP000192223 UP000076407 UP000075903 UP000079169 UP000007062 UP000008792 UP000001070 UP000075880 UP000075884 UP000009192 UP000075883 UP000015103 UP000075886 UP000075920 UP000075881 UP000076408 UP000075900 UP000000305 UP000001819 UP000007798 UP000268350 UP000069272 UP000192221 UP000000304 UP000001292 UP000000803 UP000007801 UP000008711 UP000002282 UP000075882 UP000076858 UP000019118 UP000030742
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A194QT90
A0A194QAZ4
A0A2A4J296
A0A212FFE0
A0A2J7QD95
A0A1Y1N3N7
+ More
W8ATJ8 D6WE38 A0A0T6BCT8 A0A1L8EFU0 A0A336MQ22 A0A1L8EFE2 A0A1S4FYZ9 A0A1L8EFH7 Q16JK3 A0A1L8EFE4 A0A1L8EFL8 U5EXR4 T1PIZ3 A0A1I8N4W1 A0A0L0C0S7 A0A1I8NV62 A0A0K8VHX8 A0A1Q3G301 A0A0K8UME6 A0A0A1XH39 A0A0A1X4A1 A0A0M4EV67 W5JGJ0 A0A1B6JVR4 A0A1B6FCW5 A0A1B6DK71 A0A084VNX5 A0A182IAN3 A0A1W4XD41 A0A2P2I6R7 A0A182XBV1 A0A182UUX2 A0A1S4ECY5 Q7QF54 B4M0B9 B4JRQ6 A0A0P4VRA8 A0A182IKW5 A0A182NC31 B4K746 A0A182MJD4 T1HPI4 T1D6C8 A0A182QIM8 A0A182VV65 A0A182K5P8 A0A182YR78 A0A0A9Z2Z0 A0A0A9YX13 A0A182RM61 E9FSV2 B5DVK2 B4NJQ4 A0A3B0JIF3 A0A182F8E4 A0A224XGX1 A0A1W4UMT2 A0A023F2F4 B4QSA1 B4HGX0 Q9VG64 B3M2R9 B3P4B1 B4PPA7 A0A182LM32 A0A0P5G8L1 A0A0P5S9Y5 A0A0P5RY90 A0A0P5LAZ5 A0A0P5N0F0 A0A0P5PTC8 A0A162D8Z8 A0A0P5K4Y7 A0A0P5LHK0 A0A0P5K3P3 A0A023EQW2 A0A0P5IVW9 A0A0P5P1T4 A0A0P5G7R1 J3JTV8 A0A0N8BRR6 A0A0P6ERD2 A0A0N8BQQ4 N6TWH6 A0A0P4Y8Y4 A0A0P5BCL7 A0A293N3C7 A0A0P5VMA1 A0A0P5NX51 A0A0P5Z9B7 A0A0P4Z7K9 A0A0P5GSV1 A0A0P5ZFL4 U4UBS0
W8ATJ8 D6WE38 A0A0T6BCT8 A0A1L8EFU0 A0A336MQ22 A0A1L8EFE2 A0A1S4FYZ9 A0A1L8EFH7 Q16JK3 A0A1L8EFE4 A0A1L8EFL8 U5EXR4 T1PIZ3 A0A1I8N4W1 A0A0L0C0S7 A0A1I8NV62 A0A0K8VHX8 A0A1Q3G301 A0A0K8UME6 A0A0A1XH39 A0A0A1X4A1 A0A0M4EV67 W5JGJ0 A0A1B6JVR4 A0A1B6FCW5 A0A1B6DK71 A0A084VNX5 A0A182IAN3 A0A1W4XD41 A0A2P2I6R7 A0A182XBV1 A0A182UUX2 A0A1S4ECY5 Q7QF54 B4M0B9 B4JRQ6 A0A0P4VRA8 A0A182IKW5 A0A182NC31 B4K746 A0A182MJD4 T1HPI4 T1D6C8 A0A182QIM8 A0A182VV65 A0A182K5P8 A0A182YR78 A0A0A9Z2Z0 A0A0A9YX13 A0A182RM61 E9FSV2 B5DVK2 B4NJQ4 A0A3B0JIF3 A0A182F8E4 A0A224XGX1 A0A1W4UMT2 A0A023F2F4 B4QSA1 B4HGX0 Q9VG64 B3M2R9 B3P4B1 B4PPA7 A0A182LM32 A0A0P5G8L1 A0A0P5S9Y5 A0A0P5RY90 A0A0P5LAZ5 A0A0P5N0F0 A0A0P5PTC8 A0A162D8Z8 A0A0P5K4Y7 A0A0P5LHK0 A0A0P5K3P3 A0A023EQW2 A0A0P5IVW9 A0A0P5P1T4 A0A0P5G7R1 J3JTV8 A0A0N8BRR6 A0A0P6ERD2 A0A0N8BQQ4 N6TWH6 A0A0P4Y8Y4 A0A0P5BCL7 A0A293N3C7 A0A0P5VMA1 A0A0P5NX51 A0A0P5Z9B7 A0A0P4Z7K9 A0A0P5GSV1 A0A0P5ZFL4 U4UBS0
Ontologies
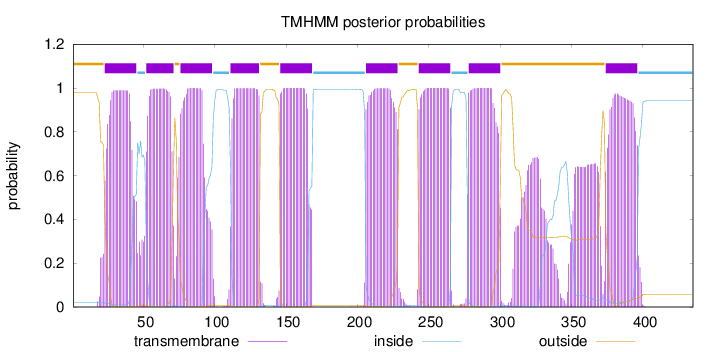
Topology
Length:
435
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
221.09002
Exp number, first 60 AAs:
30.83796
Total prob of N-in:
0.02122
POSSIBLE N-term signal
sequence
outside
1 - 22
TMhelix
23 - 45
inside
46 - 51
TMhelix
52 - 71
outside
72 - 75
TMhelix
76 - 98
inside
99 - 110
TMhelix
111 - 131
outside
132 - 145
TMhelix
146 - 168
inside
169 - 205
TMhelix
206 - 228
outside
229 - 242
TMhelix
243 - 265
inside
266 - 277
TMhelix
278 - 300
outside
301 - 373
TMhelix
374 - 396
inside
397 - 435
Population Genetic Test Statistics
Pi
215.115233
Theta
20.181502
Tajima's D
0.612732
CLR
0.555487
CSRT
0.555922203889806
Interpretation
Uncertain
