Gene
KWMTBOMO10223
Pre Gene Modal
BGIBMGA014441
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_rho_GTPase-activating_protein_44_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.212
Sequence
CDS
ATGAAGAAACAATTTTATAGAGTGAAACAACTTGCAGACCAAACGTTTTCAAGATCAGGAAAAGGAGAGGCCCTAACAGATGAACTACATACTGCTGACAGGAAAGTGGAAAACTTGCGAAATGCTTTACAGCTTGTTAGTAAAAGGCTTTCAACAGGTGCCGGAAATACACAAGGTCAAGATCCAGCTGCCAGAGAGAAAAGACTCAAAAAGCTTCCAGAGTATTTATTGGGCTTGTCCTTGTGTGAAGCTAGCAACTTTGATGATGATGACAGTATTTTAAAATTCATATTATATGAATGTGGTAAAACTGAGAAGTTCTTGGCTAACGAAATAGCTGAACACGAGTTAAAAGTGGAACAGTTGGTATGTGCACCTTTAACAGTAATCAGTGATCATGATCTACCAGCAATAATGAAAGCCAAAAAACAACTCAGCAGGCTAATTAGTGAGAAAGAGTCTGCAGCAAGTAGATACAATCATTTAGATAAACAAAAAGAGGAAAATCCCACAAAACTAGCAGCAGCCCGAGAGGAACTTGAAGAAGCTGGCACAAAAGTAGAGTCTGCACGAGATGCTCTTGCTGCGGATATGTTTGCCCTTGTAGCCAAAGAAGCTCAGCTTGCTCACACTCTACTGCAATATGTTAAATTACAGAGAGCTTACCATGAATCTGCCCTACACTCACTACAAGATACAGTACCAGAGTTGGAAAAGTTTATCAACGATAGTTCTGTGAAACCAGTTTTTGGATATCCATTGGAGGAACACTTGCGGGTTACTGGGCGCACCATTGCTTTTCCTATCGAGCTATGCGTCTGCACATTGCATGAATTGGCATTACATGAGGAGGGACTCTTCAGAATAGCTGGTGCAACATCAAAAGTGAGAAGAATGAAGCTTTCACTCGATGCGGGGTTATTCAGTGTGCCTTTACCACCGGATTACAGAGATATGCACGTTGTGGCGTCAGTACTCAAGTCATACTTGCGTGAACTGCCTGAGCCTCTACTGACATATAGGCTTTATGAGAATTTTATGAATGCGTCACGACAACCGACAGAACAGGCCAGACTTAACGCCCTATGGGAGTGTATTCATTTACTGCCCGATGCTAACTACCAAAACCTTAGATACCTAATAAAATTTTTGTCCGCTTTGACACAAAACCAAAATAAGAACAAAATGACGCCTTCAAATCTGGCCATCGTGGTGGCTCCCAACTTATTGTGGGCCGCTGATGAGAGCACTTTCGATATGAATATCACAACAGCTGTCAACTGCGTCGTGGAGCTCCTAATTAAACACGCAGACTGGTTTTTCAAGGACGAAGTTAACTTTTTCATTTCATTCACCAAAGAAGATTTGCTACCAGGCCTGTACCAAGACGGTTTCACCCCTAATTTCGGCCACGGATACAATCAAACCGCCGCCCTCGCTCAAGATCAATCTAACGGCGAATACAGCAGCATGAGCAAATCGATGTTCGATTATAACCACCACTCGGAACAGGTCAAGTACAGTCAAGTCAAGGTCGCGGCCGACGGTCCCGGCCGACATTCACGGAGCAACAGTCACGACACCAGCCTCATCTTATTAGAAAGTGACATCAAGAAGGCACAGTCCAATAGTTCCCTTTCCGACCAATCCAGCCCGCCCCACGGAAGTCCCAAACCAATAACGAGGCGGAAAAACAAACCTCTCGCTCCGGTCCCGCCAAATTTTACTCCAGACAAAAACAAAAAAACGGATACTCAAACGCAAACAACAGCCCAAACTCAAGCCGAAACTACGAGAGAGATACAGCCCGCAGCGAAAACCGAAGTCGAGAAACCGGCCAAGCCACCGAGACCAGTCATATCCGACACCGGTAAGGTGATCACTGGGGTGCAGACGATCAATCGGTCCACATATCGGCAGAACAAGGCGCTCAAGGAGGAGGCCGCCCGCGAACTCGTGTCAGATGCGCGGCGACAGAGTCTCGAGCTAGAGCGAGACGGAACGGAGCTCTCGCGAGCCGACTGCACCCTTGTCGTGCGTAACCTCACCGCGCACACCGGGGTCGTCGGCCGTATGAAGGTGGTCGGAGACGCCCGCTCCCCCCTCCTTGCTGGCAAGCAGCCCCCGCCTCGACCCGTAGCGGCACCGCGCACTCTCGTACCTGTAGCGACGGACCTCGACGCAGACGTCACCCTGCGCAACAAGCCTGCCGTGCCCGAGCGGCCGGCCACACTGCGGCCGCAAAGCTTCCACGCCCCTCGGGGCTCTTCTTCCTCTGACGTGGAAGTCGCGCCGACCGTGCTCGAGCGGACGCACATCTACACAGTGGACAAACAACAACCGACTGTGATCCAGGTGGGCGGCGGCGACGAGGCTCCGCGGGCGACTGTGCAGCGCACGCACAGTTCCGGCGGTAAGGACGCAGAGCTCGACGAGGGCAAGGGTCTGAGTGCGAGCCGGCAGCTGTCGCAGTCGGAAGGCAGTATCGCCGAGTCTCCGTCGCCGCGCGCACGCCCGGCGCGGCCGCAGGTGCCAGCGCCACCGCCGCCGGTGTCCGTCTCCTCGCCGGAGTCCGCCCCGCCGGCGTCCCTCTCGCAGCCGGAGTCACTCCCACCGGCGGCGTCGGTTCCGCTGCAGGAGAGCACAAACTTGTAG
Protein
MKKQFYRVKQLADQTFSRSGKGEALTDELHTADRKVENLRNALQLVSKRLSTGAGNTQGQDPAAREKRLKKLPEYLLGLSLCEASNFDDDDSILKFILYECGKTEKFLANEIAEHELKVEQLVCAPLTVISDHDLPAIMKAKKQLSRLISEKESAASRYNHLDKQKEENPTKLAAAREELEEAGTKVESARDALAADMFALVAKEAQLAHTLLQYVKLQRAYHESALHSLQDTVPELEKFINDSSVKPVFGYPLEEHLRVTGRTIAFPIELCVCTLHELALHEEGLFRIAGATSKVRRMKLSLDAGLFSVPLPPDYRDMHVVASVLKSYLRELPEPLLTYRLYENFMNASRQPTEQARLNALWECIHLLPDANYQNLRYLIKFLSALTQNQNKNKMTPSNLAIVVAPNLLWAADESTFDMNITTAVNCVVELLIKHADWFFKDEVNFFISFTKEDLLPGLYQDGFTPNFGHGYNQTAALAQDQSNGEYSSMSKSMFDYNHHSEQVKYSQVKVAADGPGRHSRSNSHDTSLILLESDIKKAQSNSSLSDQSSPPHGSPKPITRRKNKPLAPVPPNFTPDKNKKTDTQTQTTAQTQAETTREIQPAAKTEVEKPAKPPRPVISDTGKVITGVQTINRSTYRQNKALKEEAARELVSDARRQSLELERDGTELSRADCTLVVRNLTAHTGVVGRMKVVGDARSPLLAGKQPPPRPVAAPRTLVPVATDLDADVTLRNKPAVPERPATLRPQSFHAPRGSSSSDVEVAPTVLERTHIYTVDKQQPTVIQVGGGDEAPRATVQRTHSSGGKDAELDEGKGLSASRQLSQSEGSIAESPSPRARPARPQVPAPPPPVSVSSPESAPPASLSQPESLPPAASVPLQESTNL
Summary
Uniprot
A0A3S2LU45
A0A2H1VT66
A0A194QB73
A0A212FFE5
A0A194QXB9
A0A2A4JA08
+ More
A0A1E1WG87 A0A1E1WNA5 A0A0L7LS79 A0A2P8YZS9 A0A2J7QD87 A0A2J7QD90 A0A1L8DM05 A0A1L8DLU5 A0A1L8DLW2 A0A0N8D2K1 A0A0P5M3M8 A0A0P5E3B0 A0A0P5VZK0 A0A0P5GBG8 A0A0P5NVZ3 A0A0P5NGF1 A0A0P5R0V8 A0A0P6AI47 A0A0P5RJ46 A0A0P5QR98 A0A0P6GJJ7 A0A0P6GFL7 A0A0P6F3B6 A0A0P5C9H7 A0A0P6DJJ7 A0A0P6EZ05 A0A0P6ETH6 A0A0P5L336 A0A0P5I747
A0A1E1WG87 A0A1E1WNA5 A0A0L7LS79 A0A2P8YZS9 A0A2J7QD87 A0A2J7QD90 A0A1L8DM05 A0A1L8DLU5 A0A1L8DLW2 A0A0N8D2K1 A0A0P5M3M8 A0A0P5E3B0 A0A0P5VZK0 A0A0P5GBG8 A0A0P5NVZ3 A0A0P5NGF1 A0A0P5R0V8 A0A0P6AI47 A0A0P5RJ46 A0A0P5QR98 A0A0P6GJJ7 A0A0P6GFL7 A0A0P6F3B6 A0A0P5C9H7 A0A0P6DJJ7 A0A0P6EZ05 A0A0P6ETH6 A0A0P5L336 A0A0P5I747
EMBL
RSAL01000200
RVE44460.1
ODYU01004284
SOQ43988.1
KQ459232
KPJ02712.1
+ More
AGBW02008828 OWR52452.1 KQ461155 KPJ08221.1 NWSH01002300 PCG68586.1 GDQN01005065 JAT85989.1 GDQN01002713 JAT88341.1 JTDY01000251 KOB78056.1 PYGN01000263 PSN49744.1 NEVH01015826 PNF26550.1 PNF26551.1 GFDF01006643 JAV07441.1 GFDF01006641 JAV07443.1 GFDF01006642 JAV07442.1 GDIP01075039 JAM28676.1 GDIQ01161409 JAK90316.1 GDIP01152232 JAJ71170.1 GDIP01092936 JAM10779.1 GDIQ01270990 GDIQ01250782 GDIQ01207137 GDIQ01199450 GDIQ01162714 GDIQ01147024 JAK00943.1 GDIQ01207138 GDIQ01199449 GDIQ01169501 GDIQ01165608 GDIQ01162713 GDIQ01147025 GDIQ01099584 GDIQ01097987 GDIQ01032205 JAL04701.1 GDIQ01153215 JAK98510.1 GDIQ01107119 JAL44607.1 GDIP01029445 JAM74270.1 GDIQ01109320 JAL42406.1 GDIQ01111126 JAL40600.1 GDIQ01033811 JAN60926.1 GDIQ01033810 JAN60927.1 GDIQ01058144 GDIQ01055547 JAN39190.1 GDIP01189866 JAJ33536.1 GDIQ01075937 JAN18800.1 GDIQ01055495 JAN39242.1 GDIQ01058145 JAN36592.1 GDIQ01176101 JAK75624.1 GDIQ01218580 JAK33145.1
AGBW02008828 OWR52452.1 KQ461155 KPJ08221.1 NWSH01002300 PCG68586.1 GDQN01005065 JAT85989.1 GDQN01002713 JAT88341.1 JTDY01000251 KOB78056.1 PYGN01000263 PSN49744.1 NEVH01015826 PNF26550.1 PNF26551.1 GFDF01006643 JAV07441.1 GFDF01006641 JAV07443.1 GFDF01006642 JAV07442.1 GDIP01075039 JAM28676.1 GDIQ01161409 JAK90316.1 GDIP01152232 JAJ71170.1 GDIP01092936 JAM10779.1 GDIQ01270990 GDIQ01250782 GDIQ01207137 GDIQ01199450 GDIQ01162714 GDIQ01147024 JAK00943.1 GDIQ01207138 GDIQ01199449 GDIQ01169501 GDIQ01165608 GDIQ01162713 GDIQ01147025 GDIQ01099584 GDIQ01097987 GDIQ01032205 JAL04701.1 GDIQ01153215 JAK98510.1 GDIQ01107119 JAL44607.1 GDIP01029445 JAM74270.1 GDIQ01109320 JAL42406.1 GDIQ01111126 JAL40600.1 GDIQ01033811 JAN60926.1 GDIQ01033810 JAN60927.1 GDIQ01058144 GDIQ01055547 JAN39190.1 GDIP01189866 JAJ33536.1 GDIQ01075937 JAN18800.1 GDIQ01055495 JAN39242.1 GDIQ01058145 JAN36592.1 GDIQ01176101 JAK75624.1 GDIQ01218580 JAK33145.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A3S2LU45
A0A2H1VT66
A0A194QB73
A0A212FFE5
A0A194QXB9
A0A2A4JA08
+ More
A0A1E1WG87 A0A1E1WNA5 A0A0L7LS79 A0A2P8YZS9 A0A2J7QD87 A0A2J7QD90 A0A1L8DM05 A0A1L8DLU5 A0A1L8DLW2 A0A0N8D2K1 A0A0P5M3M8 A0A0P5E3B0 A0A0P5VZK0 A0A0P5GBG8 A0A0P5NVZ3 A0A0P5NGF1 A0A0P5R0V8 A0A0P6AI47 A0A0P5RJ46 A0A0P5QR98 A0A0P6GJJ7 A0A0P6GFL7 A0A0P6F3B6 A0A0P5C9H7 A0A0P6DJJ7 A0A0P6EZ05 A0A0P6ETH6 A0A0P5L336 A0A0P5I747
A0A1E1WG87 A0A1E1WNA5 A0A0L7LS79 A0A2P8YZS9 A0A2J7QD87 A0A2J7QD90 A0A1L8DM05 A0A1L8DLU5 A0A1L8DLW2 A0A0N8D2K1 A0A0P5M3M8 A0A0P5E3B0 A0A0P5VZK0 A0A0P5GBG8 A0A0P5NVZ3 A0A0P5NGF1 A0A0P5R0V8 A0A0P6AI47 A0A0P5RJ46 A0A0P5QR98 A0A0P6GJJ7 A0A0P6GFL7 A0A0P6F3B6 A0A0P5C9H7 A0A0P6DJJ7 A0A0P6EZ05 A0A0P6ETH6 A0A0P5L336 A0A0P5I747
PDB
3IUG
E-value=4.6803e-23,
Score=270
Ontologies
GO
Topology
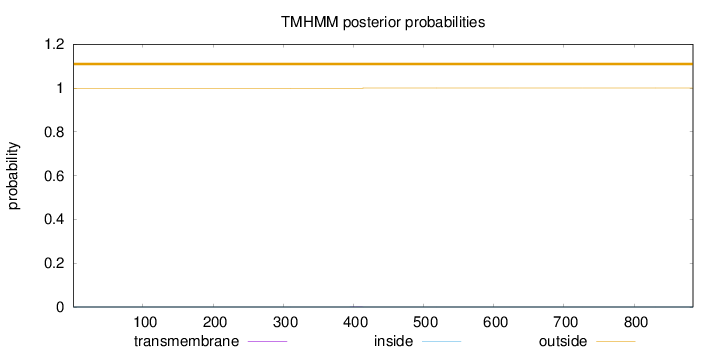
Length:
884
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00736999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00056
outside
1 - 884
Population Genetic Test Statistics
Pi
173.523657
Theta
176.456785
Tajima's D
-0.435117
CLR
2.131363
CSRT
0.261536923153842
Interpretation
Uncertain
