Gene
KWMTBOMO10222
Pre Gene Modal
BGIBMGA014443
Annotation
PREDICTED:_tyrosine--tRNA_ligase?_mitochondrial_[Papilio_machaon]
Full name
Tyrosine--tRNA ligase
+ More
Tyrosine--tRNA ligase, mitochondrial
Alkaline phosphatase
Tyrosine--tRNA ligase, mitochondrial
Alkaline phosphatase
Alternative Name
Tyrosyl-tRNA synthetase
Mitochondrial tyrosyl-tRNA synthetase
Mitochondrial tyrosyl-tRNA synthetase
Location in the cell
Mitochondrial Reliability : 1.779
Sequence
CDS
ATGTATCAAGATTTGTTCCCGAACACTGCGGGAAAAGAAATATTAAATTTGTTGAACAAAGCCCCACAATGTGTTTATGCTGGTTTTGATCCAACAGCTGATAGCCTTCATGTTGGGAATCTACTGGTAATAATAAATCTACTCCATTGGTACAGAGGAGGACACAAAGTCATTGCATTGTTAGGGGGGGCAACTGGGCATATTGGTGACCCGAGTGGCAGAAACACTGATCGTGCAGCATTACAAAAAGAAGTTATAGAAAAAAATATAAAATGTATCAGGAACAACTTGGAAACTATATTTGAAAATCACAATAAATACATCTCATCCAAAGATGATGGACCACCACAACCACTAAGAATAGTCAATAATGAAGAATGGTATAGAAAAATAGATTCAATACAGTTTGTAAGCGAAATAGGTCGTAATTTTCGGATGGGCACAATGCTGTTGAAACACAGTGTTCAATCTAGAATCAACTCTGAAGTTGGCATGAGTTTCACCGAATTTAGCTATCAGATATTTCAGTCATATGATTGGTTACATTTACTAAAAGAATATGACTGTAGGTTTCAGGTAGGTGGCAGTGATCAGATGGGTAATATCACAGCTGGCCATGAATTAATAAGTAGAACTGCAAAGACAGATGTTTATGGTTTAACATTGCCATTAGTAACAACAGATGAAGGGGATAAGTTTGGAAAGTCTGCAGGAAATGCATTGTGGTTGGATTCAGCAAAAACAAGTCCATTTAGTTTGTATCAATATTTCATTCGAACCAAAGATGCTGATGTAGAGCGACTGTTGAAGCTATTTACTTTCTACAGCTTAGGAGAGATCAGAGATATCATGTATAAACATGAACAGCATCCGGATCAAAGGTATCCACAGCAGTGTCTGGCTGAGCAGCTAACGACTCTGGTGCATGGAAAGGATGGATTAGCCCAAGCACAAAGAGCAACATCTGCTATTTACAGTAAGGATATTAAATCATTAGTCTCGTTGAAGAGTGATGAGTTGGAAAAAGTTTTCGAAGGTGCTCCCGTTACTAGTTTATTATTATCACCTGGAATAACAGTTTTAGAACTTGGAATGAAAGCTAAATGCTTCCCGACTGAGAATGATGCCATGAGAATTATAGAAGCCGGTGGATTCTACATCAATCATCAGAAAATAAAGAAAATTGATGAGGTCATAACACAATCAGCACACATACTACCAAATCTAATATCACTCTTAAGGACTGGGAAACGTAATTATTATATTGTGAAATGGCAAACATAA
Protein
MYQDLFPNTAGKEILNLLNKAPQCVYAGFDPTADSLHVGNLLVIINLLHWYRGGHKVIALLGGATGHIGDPSGRNTDRAALQKEVIEKNIKCIRNNLETIFENHNKYISSKDDGPPQPLRIVNNEEWYRKIDSIQFVSEIGRNFRMGTMLLKHSVQSRINSEVGMSFTEFSYQIFQSYDWLHLLKEYDCRFQVGGSDQMGNITAGHELISRTAKTDVYGLTLPLVTTDEGDKFGKSAGNALWLDSAKTSPFSLYQYFIRTKDADVERLLKLFTFYSLGEIRDIMYKHEQHPDQRYPQQCLAEQLTTLVHGKDGLAQAQRATSAIYSKDIKSLVSLKSDELEKVFEGAPVTSLLLSPGITVLELGMKAKCFPTENDAMRIIEAGGFYINHQKIKKIDEVITQSAHILPNLISLLRTGKRNYYIVKWQT
Summary
Description
Catalyzes the attachment of tyrosine to tRNA(Tyr) in a two-step reaction: tyrosine is first activated by ATP to form Tyr-AMP and then transferred to the acceptor end of tRNA(Tyr).
Catalytic Activity
ATP + L-tyrosine + tRNA(Tyr) = AMP + diphosphate + H(+) + L-tyrosyl-tRNA(Tyr)
a phosphate monoester + H2O = an alcohol + phosphate
a phosphate monoester + H2O = an alcohol + phosphate
Subunit
Homodimer.
Similarity
Belongs to the class-I aminoacyl-tRNA synthetase family.
Belongs to the alkaline phosphatase family.
Belongs to the alkaline phosphatase family.
Keywords
Aminoacyl-tRNA synthetase
ATP-binding
Complete proteome
Ligase
Mitochondrion
Nucleotide-binding
Protein biosynthesis
Reference proteome
Transit peptide
Feature
chain Tyrosine--tRNA ligase, mitochondrial
Uniprot
H9JY25
A0A2H1VT49
A0A2A4J8Z0
A0A194QGZ0
A0A212FFF8
A0A194QRV7
+ More
A0A0L7LS86 A0A195CLP7 A0A2S2N9A3 J9JRX2 A0A023F3D0 F4W6P1 A0A151HZV4 A0A151XAQ3 A0A158P2I1 A0A2J7RIN6 A0A195EQN4 A0A151J8N6 Q16SL3 A0A3B0JTV9 A0A1I8MD56 A0A0K8T3L0 T1HKY5 A0A0A9Y2U1 E2B667 Q28Z21 A0A1B0DBD8 A0A182H500 A0A1Y1M479 B4J4D4 B4KTJ0 B4PBV0 A0A1B6D968 B4LN55 B3MC25 A0A1B6C6N0 A0A0J9RLZ0 B4IH83 B3NQY8 Q9W107 K7ITI4 A0A1W4UZI1 A0A232EI78 A0A0M3QV72 A0A1B0FDZ9 E2AGL3 A0A0K8VEW0 E9J3V7 A0A1A9ZE36 A0A1A9VNJ8 A0A1I8NNG3 B4MP96 D3TL61 A0A0L0CB85 W8C8K7 A0A1B6IT28 A0A034VI70 A0A0A1XNE5 A0A1A9WQR0 E0VHG3 N6U5M8 A0A1B6F2B0 A0A1B6CPM2 A0A336MKG6 A0A336MFZ3 A0A1B6CXB4 A0A0C9QM19 A0A1B0AZK7 B4QCK0 A0A1B0CT28 A0A182PM56 A0A139WDX0 A0A067R4R7 A0A0L7R0G5 E9FWE8 A0A182VVI3 A0A182I9Q2 A0A182X584 A0A0P5XC04 A0A182V3U7 Q7QEJ0 A0A0P5XC23 U5ER20 A0A0P5KB86 A0A0P5CNU8 A0A182R226 A0A1A9YMY8 A0A182UL79 A0A1J1J372 A0A154NXY0 A0A182QLZ4 A0A182N144 A0A2P8XID7 A0A182J403 A0A0J7KLY6 A0A182FIV6 A0A0P6C717 A0A182MGK4 A0A3R7SLX7 A0A2M4AVS9
A0A0L7LS86 A0A195CLP7 A0A2S2N9A3 J9JRX2 A0A023F3D0 F4W6P1 A0A151HZV4 A0A151XAQ3 A0A158P2I1 A0A2J7RIN6 A0A195EQN4 A0A151J8N6 Q16SL3 A0A3B0JTV9 A0A1I8MD56 A0A0K8T3L0 T1HKY5 A0A0A9Y2U1 E2B667 Q28Z21 A0A1B0DBD8 A0A182H500 A0A1Y1M479 B4J4D4 B4KTJ0 B4PBV0 A0A1B6D968 B4LN55 B3MC25 A0A1B6C6N0 A0A0J9RLZ0 B4IH83 B3NQY8 Q9W107 K7ITI4 A0A1W4UZI1 A0A232EI78 A0A0M3QV72 A0A1B0FDZ9 E2AGL3 A0A0K8VEW0 E9J3V7 A0A1A9ZE36 A0A1A9VNJ8 A0A1I8NNG3 B4MP96 D3TL61 A0A0L0CB85 W8C8K7 A0A1B6IT28 A0A034VI70 A0A0A1XNE5 A0A1A9WQR0 E0VHG3 N6U5M8 A0A1B6F2B0 A0A1B6CPM2 A0A336MKG6 A0A336MFZ3 A0A1B6CXB4 A0A0C9QM19 A0A1B0AZK7 B4QCK0 A0A1B0CT28 A0A182PM56 A0A139WDX0 A0A067R4R7 A0A0L7R0G5 E9FWE8 A0A182VVI3 A0A182I9Q2 A0A182X584 A0A0P5XC04 A0A182V3U7 Q7QEJ0 A0A0P5XC23 U5ER20 A0A0P5KB86 A0A0P5CNU8 A0A182R226 A0A1A9YMY8 A0A182UL79 A0A1J1J372 A0A154NXY0 A0A182QLZ4 A0A182N144 A0A2P8XID7 A0A182J403 A0A0J7KLY6 A0A182FIV6 A0A0P6C717 A0A182MGK4 A0A3R7SLX7 A0A2M4AVS9
EC Number
6.1.1.1
3.1.3.1
3.1.3.1
Pubmed
19121390
26354079
22118469
26227816
25474469
21719571
+ More
21347285 17510324 25315136 25401762 26823975 20798317 15632085 17994087 26483478 28004739 17550304 22936249 18057021 10731132 12537572 12537569 20075255 28648823 21282665 20353571 26108605 24495485 25348373 25830018 20566863 23537049 18362917 19820115 24845553 21292972 12364791 14747013 17210077 29403074
21347285 17510324 25315136 25401762 26823975 20798317 15632085 17994087 26483478 28004739 17550304 22936249 18057021 10731132 12537572 12537569 20075255 28648823 21282665 20353571 26108605 24495485 25348373 25830018 20566863 23537049 18362917 19820115 24845553 21292972 12364791 14747013 17210077 29403074
EMBL
BABH01041812
ODYU01004284
SOQ43989.1
NWSH01002300
PCG68587.1
KQ459232
+ More
KPJ02711.1 AGBW02008828 OWR52453.1 KQ461155 KPJ08222.1 JTDY01000251 KOB78061.1 KQ977595 KYN01625.1 GGMR01000707 MBY13326.1 ABLF02012310 GBBI01002924 JAC15788.1 GL887707 EGI70117.1 KQ976675 KYM78226.1 KQ982335 KYQ57446.1 ADTU01006781 NEVH01003493 PNF40699.1 KQ982021 KYN30486.1 KQ979492 KYN21217.1 CH477672 EAT37451.1 OUUW01000001 SPP74518.1 GBRD01005712 JAG60109.1 ACPB03020517 GBHO01017135 GDHC01005149 JAG26469.1 JAQ13480.1 GL445930 EFN88760.1 CM000071 EAL25794.1 AJVK01004854 AJVK01004855 AJVK01004856 JXUM01110506 KQ565419 KXJ71001.1 GEZM01043894 JAV78676.1 CH916367 EDW01616.1 CH933808 EDW08551.1 CM000158 EDW92604.1 GEDC01015057 JAS22241.1 CH940648 EDW60059.1 CH902619 EDV37212.1 GEDC01028234 JAS09064.1 CM002911 KMY96434.1 CH480838 EDW49259.1 CH954179 EDV57071.1 AE013599 AY060975 AY113395 BT030810 AAZX01003331 NNAY01004350 OXU18021.1 CP012524 ALC41909.1 CCAG010004895 GL439322 EFN67433.1 GDHF01029121 GDHF01024460 GDHF01014825 GDHF01006952 JAI23193.1 JAI27854.1 JAI37489.1 JAI45362.1 GL768069 EFZ12549.1 CH963849 EDW73935.1 EZ422161 ADD18356.1 JRES01000655 KNC29516.1 GAMC01002969 JAC03587.1 GECU01017612 JAS90094.1 GAKP01015946 GAKP01015944 JAC43006.1 GBXI01001478 JAD12814.1 DS235170 EEB12819.1 APGK01038980 KB740966 KB631578 ENN76915.1 ERL84399.1 GECZ01025455 JAS44314.1 GEDC01022055 JAS15243.1 UFQT01001201 SSX29479.1 UFQT01001175 SSX29322.1 GEDC01019188 JAS18110.1 GBYB01004584 JAG74351.1 JXJN01006356 CM000362 EDX08616.1 AJWK01026956 AJWK01026957 AJWK01026958 AJWK01026959 KQ971357 KYB26031.1 KK852980 KDR12986.1 KQ414670 KOC64311.1 GL732526 EFX88442.1 APCN01001016 GDIP01074309 JAM29406.1 AAAB01008846 EAA06615.3 GDIP01074308 JAM29407.1 GANO01003006 JAB56865.1 GDIP01233776 GDIP01164578 GDIQ01241443 GDIQ01186812 GDIQ01186811 GDIP01074310 GDIQ01054665 JAI89625.1 JAJ58824.1 JAK64913.1 GDIP01168935 JAJ54467.1 CVRI01000064 CRL05321.1 KQ434781 KZC04473.1 AXCN02002167 PYGN01002030 PSN31760.1 LBMM01005582 KMQ91403.1 GDIP01007922 JAM95793.1 AXCM01000981 QCYY01002955 ROT66341.1 GGFK01011553 MBW44874.1
KPJ02711.1 AGBW02008828 OWR52453.1 KQ461155 KPJ08222.1 JTDY01000251 KOB78061.1 KQ977595 KYN01625.1 GGMR01000707 MBY13326.1 ABLF02012310 GBBI01002924 JAC15788.1 GL887707 EGI70117.1 KQ976675 KYM78226.1 KQ982335 KYQ57446.1 ADTU01006781 NEVH01003493 PNF40699.1 KQ982021 KYN30486.1 KQ979492 KYN21217.1 CH477672 EAT37451.1 OUUW01000001 SPP74518.1 GBRD01005712 JAG60109.1 ACPB03020517 GBHO01017135 GDHC01005149 JAG26469.1 JAQ13480.1 GL445930 EFN88760.1 CM000071 EAL25794.1 AJVK01004854 AJVK01004855 AJVK01004856 JXUM01110506 KQ565419 KXJ71001.1 GEZM01043894 JAV78676.1 CH916367 EDW01616.1 CH933808 EDW08551.1 CM000158 EDW92604.1 GEDC01015057 JAS22241.1 CH940648 EDW60059.1 CH902619 EDV37212.1 GEDC01028234 JAS09064.1 CM002911 KMY96434.1 CH480838 EDW49259.1 CH954179 EDV57071.1 AE013599 AY060975 AY113395 BT030810 AAZX01003331 NNAY01004350 OXU18021.1 CP012524 ALC41909.1 CCAG010004895 GL439322 EFN67433.1 GDHF01029121 GDHF01024460 GDHF01014825 GDHF01006952 JAI23193.1 JAI27854.1 JAI37489.1 JAI45362.1 GL768069 EFZ12549.1 CH963849 EDW73935.1 EZ422161 ADD18356.1 JRES01000655 KNC29516.1 GAMC01002969 JAC03587.1 GECU01017612 JAS90094.1 GAKP01015946 GAKP01015944 JAC43006.1 GBXI01001478 JAD12814.1 DS235170 EEB12819.1 APGK01038980 KB740966 KB631578 ENN76915.1 ERL84399.1 GECZ01025455 JAS44314.1 GEDC01022055 JAS15243.1 UFQT01001201 SSX29479.1 UFQT01001175 SSX29322.1 GEDC01019188 JAS18110.1 GBYB01004584 JAG74351.1 JXJN01006356 CM000362 EDX08616.1 AJWK01026956 AJWK01026957 AJWK01026958 AJWK01026959 KQ971357 KYB26031.1 KK852980 KDR12986.1 KQ414670 KOC64311.1 GL732526 EFX88442.1 APCN01001016 GDIP01074309 JAM29406.1 AAAB01008846 EAA06615.3 GDIP01074308 JAM29407.1 GANO01003006 JAB56865.1 GDIP01233776 GDIP01164578 GDIQ01241443 GDIQ01186812 GDIQ01186811 GDIP01074310 GDIQ01054665 JAI89625.1 JAJ58824.1 JAK64913.1 GDIP01168935 JAJ54467.1 CVRI01000064 CRL05321.1 KQ434781 KZC04473.1 AXCN02002167 PYGN01002030 PSN31760.1 LBMM01005582 KMQ91403.1 GDIP01007922 JAM95793.1 AXCM01000981 QCYY01002955 ROT66341.1 GGFK01011553 MBW44874.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000037510
+ More
UP000078542 UP000007819 UP000007755 UP000078540 UP000075809 UP000005205 UP000235965 UP000078541 UP000078492 UP000008820 UP000268350 UP000095301 UP000015103 UP000008237 UP000001819 UP000092462 UP000069940 UP000249989 UP000001070 UP000009192 UP000002282 UP000008792 UP000007801 UP000001292 UP000008711 UP000000803 UP000002358 UP000192221 UP000215335 UP000092553 UP000092444 UP000000311 UP000092445 UP000078200 UP000095300 UP000007798 UP000037069 UP000091820 UP000009046 UP000019118 UP000030742 UP000092460 UP000000304 UP000092461 UP000075885 UP000007266 UP000027135 UP000053825 UP000000305 UP000075920 UP000075840 UP000076407 UP000075903 UP000007062 UP000075900 UP000092443 UP000075902 UP000183832 UP000076502 UP000075886 UP000075884 UP000245037 UP000075880 UP000036403 UP000069272 UP000075883 UP000283509
UP000078542 UP000007819 UP000007755 UP000078540 UP000075809 UP000005205 UP000235965 UP000078541 UP000078492 UP000008820 UP000268350 UP000095301 UP000015103 UP000008237 UP000001819 UP000092462 UP000069940 UP000249989 UP000001070 UP000009192 UP000002282 UP000008792 UP000007801 UP000001292 UP000008711 UP000000803 UP000002358 UP000192221 UP000215335 UP000092553 UP000092444 UP000000311 UP000092445 UP000078200 UP000095300 UP000007798 UP000037069 UP000091820 UP000009046 UP000019118 UP000030742 UP000092460 UP000000304 UP000092461 UP000075885 UP000007266 UP000027135 UP000053825 UP000000305 UP000075920 UP000075840 UP000076407 UP000075903 UP000007062 UP000075900 UP000092443 UP000075902 UP000183832 UP000076502 UP000075886 UP000075884 UP000245037 UP000075880 UP000036403 UP000069272 UP000075883 UP000283509
Interpro
IPR036986
S4_RNA-bd_sf
+ More
IPR001412 aa-tRNA-synth_I_CS
IPR014729 Rossmann-like_a/b/a_fold
IPR024088 Tyr-tRNA-ligase_bac-type
IPR002307 Tyr-tRNA-ligase
IPR002305 aa-tRNA-synth_Ic
IPR024107 Tyr-tRNA-ligase_bac_1
IPR002942 S4_RNA-bd
IPR017452 GPCR_Rhodpsn_7TM
IPR000276 GPCR_Rhodpsn
IPR018299 Alkaline_phosphatase_AS
IPR001952 Alkaline_phosphatase
IPR017850 Alkaline_phosphatase_core_sf
IPR018490 cNMP-bd-like
IPR006916 Popeye_prot
IPR036871 PX_dom_sf
IPR001683 Phox
IPR001412 aa-tRNA-synth_I_CS
IPR014729 Rossmann-like_a/b/a_fold
IPR024088 Tyr-tRNA-ligase_bac-type
IPR002307 Tyr-tRNA-ligase
IPR002305 aa-tRNA-synth_Ic
IPR024107 Tyr-tRNA-ligase_bac_1
IPR002942 S4_RNA-bd
IPR017452 GPCR_Rhodpsn_7TM
IPR000276 GPCR_Rhodpsn
IPR018299 Alkaline_phosphatase_AS
IPR001952 Alkaline_phosphatase
IPR017850 Alkaline_phosphatase_core_sf
IPR018490 cNMP-bd-like
IPR006916 Popeye_prot
IPR036871 PX_dom_sf
IPR001683 Phox
ProteinModelPortal
H9JY25
A0A2H1VT49
A0A2A4J8Z0
A0A194QGZ0
A0A212FFF8
A0A194QRV7
+ More
A0A0L7LS86 A0A195CLP7 A0A2S2N9A3 J9JRX2 A0A023F3D0 F4W6P1 A0A151HZV4 A0A151XAQ3 A0A158P2I1 A0A2J7RIN6 A0A195EQN4 A0A151J8N6 Q16SL3 A0A3B0JTV9 A0A1I8MD56 A0A0K8T3L0 T1HKY5 A0A0A9Y2U1 E2B667 Q28Z21 A0A1B0DBD8 A0A182H500 A0A1Y1M479 B4J4D4 B4KTJ0 B4PBV0 A0A1B6D968 B4LN55 B3MC25 A0A1B6C6N0 A0A0J9RLZ0 B4IH83 B3NQY8 Q9W107 K7ITI4 A0A1W4UZI1 A0A232EI78 A0A0M3QV72 A0A1B0FDZ9 E2AGL3 A0A0K8VEW0 E9J3V7 A0A1A9ZE36 A0A1A9VNJ8 A0A1I8NNG3 B4MP96 D3TL61 A0A0L0CB85 W8C8K7 A0A1B6IT28 A0A034VI70 A0A0A1XNE5 A0A1A9WQR0 E0VHG3 N6U5M8 A0A1B6F2B0 A0A1B6CPM2 A0A336MKG6 A0A336MFZ3 A0A1B6CXB4 A0A0C9QM19 A0A1B0AZK7 B4QCK0 A0A1B0CT28 A0A182PM56 A0A139WDX0 A0A067R4R7 A0A0L7R0G5 E9FWE8 A0A182VVI3 A0A182I9Q2 A0A182X584 A0A0P5XC04 A0A182V3U7 Q7QEJ0 A0A0P5XC23 U5ER20 A0A0P5KB86 A0A0P5CNU8 A0A182R226 A0A1A9YMY8 A0A182UL79 A0A1J1J372 A0A154NXY0 A0A182QLZ4 A0A182N144 A0A2P8XID7 A0A182J403 A0A0J7KLY6 A0A182FIV6 A0A0P6C717 A0A182MGK4 A0A3R7SLX7 A0A2M4AVS9
A0A0L7LS86 A0A195CLP7 A0A2S2N9A3 J9JRX2 A0A023F3D0 F4W6P1 A0A151HZV4 A0A151XAQ3 A0A158P2I1 A0A2J7RIN6 A0A195EQN4 A0A151J8N6 Q16SL3 A0A3B0JTV9 A0A1I8MD56 A0A0K8T3L0 T1HKY5 A0A0A9Y2U1 E2B667 Q28Z21 A0A1B0DBD8 A0A182H500 A0A1Y1M479 B4J4D4 B4KTJ0 B4PBV0 A0A1B6D968 B4LN55 B3MC25 A0A1B6C6N0 A0A0J9RLZ0 B4IH83 B3NQY8 Q9W107 K7ITI4 A0A1W4UZI1 A0A232EI78 A0A0M3QV72 A0A1B0FDZ9 E2AGL3 A0A0K8VEW0 E9J3V7 A0A1A9ZE36 A0A1A9VNJ8 A0A1I8NNG3 B4MP96 D3TL61 A0A0L0CB85 W8C8K7 A0A1B6IT28 A0A034VI70 A0A0A1XNE5 A0A1A9WQR0 E0VHG3 N6U5M8 A0A1B6F2B0 A0A1B6CPM2 A0A336MKG6 A0A336MFZ3 A0A1B6CXB4 A0A0C9QM19 A0A1B0AZK7 B4QCK0 A0A1B0CT28 A0A182PM56 A0A139WDX0 A0A067R4R7 A0A0L7R0G5 E9FWE8 A0A182VVI3 A0A182I9Q2 A0A182X584 A0A0P5XC04 A0A182V3U7 Q7QEJ0 A0A0P5XC23 U5ER20 A0A0P5KB86 A0A0P5CNU8 A0A182R226 A0A1A9YMY8 A0A182UL79 A0A1J1J372 A0A154NXY0 A0A182QLZ4 A0A182N144 A0A2P8XID7 A0A182J403 A0A0J7KLY6 A0A182FIV6 A0A0P6C717 A0A182MGK4 A0A3R7SLX7 A0A2M4AVS9
PDB
3ZXI
E-value=9.5713e-87,
Score=816
Ontologies
GO
PANTHER
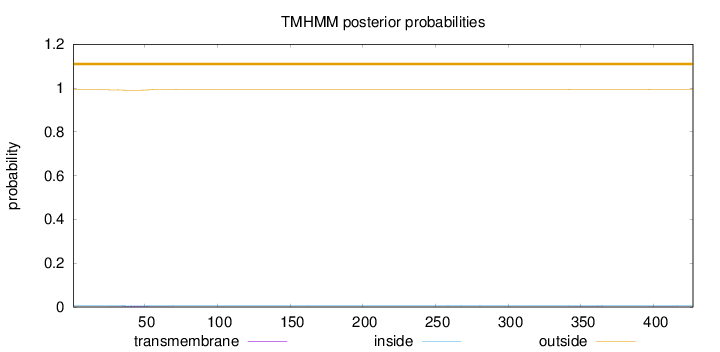
Topology
Subcellular location
Mitochondrion matrix
Length:
427
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.11911
Exp number, first 60 AAs:
0.08927
Total prob of N-in:
0.00829
outside
1 - 427
Population Genetic Test Statistics
Pi
16.77067
Theta
15.51283
Tajima's D
0.009047
CLR
2.609893
CSRT
0.373631318434078
Interpretation
Uncertain
