Gene
KWMTBOMO10221 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014442
Annotation
PREDICTED:_phosphoribosyl_pyrophosphate_synthase-associated_protein_2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.554 Nuclear Reliability : 1.682
Sequence
CDS
ATGGACGGGAACTCCACGTCGGACATTGTTATTTTGAGTGGAAACTCACATCCTGAATTAGCTGATTTGATTGCCAACCGCCTAGGTGTGCGTAAAGGAGGTTGTTCAGTATACCACAAGACTAACAGAGAGACCATTGTGGAAATTGCTGATTCAATTCGTGGCAAGAATATTTATATCATACAGACTGGAACTAAAGATGTGAACAACAACATAATGGAATTGCTTATAATGGCCTACGCTTGTAAGACTTCCTCTGCCAGTTCAATTGTTGGTGTAATTCCATACTTACCATACAGTAAACAATGCAAAATGCGGAAACGAGGATGTATAGTTTCTAAACTTTTAGCCCAAATGATGTGTAAATCAGGTCTTTCACACATTATCACTATGGATTTACACCAAAAGGAAATACAAGGATTTTTTGACTGCCCTGTTGATAATTTAAGAGCATCACCATTCTTACTTCAGTATATACAAGAAAGTGTGCAAGATTACCGTAATTCGGTTATAGTGGCCCGGAATCCTGGCTCTGCAAAGAAAGCTACTTCATATGCCGAGCGATTGAGACTAGCTATAGCTGTCATACATGGAGAACAGAAGGAAGCCGAAAGTGATGAGGTTGATGGACGGTATTCACCTCCATGTATACCCAGGTCGCGTACTATGGATGTATCTGTGGGAGTGCCGGCACATCCAGCTAAAGAAAAGCCACCGATAAATGTAGTAGGAGATGTAGGAGGACGAATTGCTATAATGGTGGACGACATGATCGACGACGTTCAATCGTTCGTGGCGGCTGCTGAAGTATTAAAAGAGTGCGGAGCCTACAAAATATATGTACTGGCCACCCACGGCCTGCTCTCGTCCGACGCTCCACGATTAATTGAGGATTCGCCCATAGACGAGGTTGTCGTCACCAACACGGTGCCTCATGAACTACAAAAGATGCAGTGCAATAAGATCAAAACTATCGATATATCGATACTGATCAGCGAAGCGATCCGGCGCATTCATAACAAAGAATCTATGTCTTATCTCTTCAAAAACGTTACTCTTGAAGATTAA
Protein
MDGNSTSDIVILSGNSHPELADLIANRLGVRKGGCSVYHKTNRETIVEIADSIRGKNIYIIQTGTKDVNNNIMELLIMAYACKTSSASSIVGVIPYLPYSKQCKMRKRGCIVSKLLAQMMCKSGLSHIITMDLHQKEIQGFFDCPVDNLRASPFLLQYIQESVQDYRNSVIVARNPGSAKKATSYAERLRLAIAVIHGEQKEAESDEVDGRYSPPCIPRSRTMDVSVGVPAHPAKEKPPINVVGDVGGRIAIMVDDMIDDVQSFVAAAEVLKECGAYKIYVLATHGLLSSDAPRLIEDSPIDEVVVTNTVPHELQKMQCNKIKTIDISILISEAIRRIHNKESMSYLFKNVTLED
Summary
Uniprot
A0A212FFD9
A0A2H1VT34
A0A194QCK1
A0A2A4J9R9
A0A2A4JA90
A0A2A4J9G1
+ More
S4NY76 A0A194QRT0 A0A0L7LRH4 A0A067QSW9 A0A1Q3FUK3 A0A1Q3F4W5 A0A2P8XLH9 A0A1L8DXV3 A0A2J7RIR1 U5EJ30 A0A023EPD2 B0WZD8 A0A1W4XDU5 A0A182Q031 A0A182Y555 A0A182M4D5 A0A182P292 A0A182VFU8 A0A182XGN9 F5HJ15 A0A182IDM3 F5HJ16 Q176H6 D6WV67 A0A1B6KQH7 A0A2M3ZI16 A0A182W461 A0A2M4AJF0 A0A2M4BRN0 W5JMP8 A0A1Y1NB29 A0A2M3Z8E2 A0A2M4AJD8 A0A2M4AJH2 A0A182FMC3 A0A2M4BRK1 A0A2M4AJD3 A0A182MZK4 A0A1B6DT63 A0A154PBZ1 A0A2A3E7S4 A0A0M8ZST7 A0A088A2Q4 N6SZF4 E0VEP9 A0A0K8TSJ4 A0A0P6BZV0 F4WFK8 A0A1I8PA62 E2AAI5 A0A026WKJ1 A0A0C9R767 A0A151HYR2 A0A158NSI1 A0A164VLK5 D3TLH8 A0A0N8API6 E9FWJ1 A0A1A9X421 A0A1B0FRE4 A0A0M4EPC1 A0A1A9XT98 R4WDK8 T1PEN3 A0A1A9V4X9 A0A1B0BEZ9 A0A1A9ZZN5 A0A1I8PA56 A0A023F9K4 A0A0P4W463 A0A1I8PA55 A0A069DTA1 R4FNN1 A0A1I8PAA7 A0A224XEU6 A0A195CS46 A0A151J440 A0A2S2PVN4 A0A1I8N3W4 A0A0L0BL65 A0A0V0G9A2 T1PP27 A0A2S2RA84 A0A0Q9XCK0 A0A0Q9XDI8 A0A0P8XU96 A0A3L8DXE8 A0A0Q9WRZ5 A0A0Q9WT90 A0A0Q9X344 A0A2H8TMG4 J9JN11 A0A2S2NL54 A0A182IJC7
S4NY76 A0A194QRT0 A0A0L7LRH4 A0A067QSW9 A0A1Q3FUK3 A0A1Q3F4W5 A0A2P8XLH9 A0A1L8DXV3 A0A2J7RIR1 U5EJ30 A0A023EPD2 B0WZD8 A0A1W4XDU5 A0A182Q031 A0A182Y555 A0A182M4D5 A0A182P292 A0A182VFU8 A0A182XGN9 F5HJ15 A0A182IDM3 F5HJ16 Q176H6 D6WV67 A0A1B6KQH7 A0A2M3ZI16 A0A182W461 A0A2M4AJF0 A0A2M4BRN0 W5JMP8 A0A1Y1NB29 A0A2M3Z8E2 A0A2M4AJD8 A0A2M4AJH2 A0A182FMC3 A0A2M4BRK1 A0A2M4AJD3 A0A182MZK4 A0A1B6DT63 A0A154PBZ1 A0A2A3E7S4 A0A0M8ZST7 A0A088A2Q4 N6SZF4 E0VEP9 A0A0K8TSJ4 A0A0P6BZV0 F4WFK8 A0A1I8PA62 E2AAI5 A0A026WKJ1 A0A0C9R767 A0A151HYR2 A0A158NSI1 A0A164VLK5 D3TLH8 A0A0N8API6 E9FWJ1 A0A1A9X421 A0A1B0FRE4 A0A0M4EPC1 A0A1A9XT98 R4WDK8 T1PEN3 A0A1A9V4X9 A0A1B0BEZ9 A0A1A9ZZN5 A0A1I8PA56 A0A023F9K4 A0A0P4W463 A0A1I8PA55 A0A069DTA1 R4FNN1 A0A1I8PAA7 A0A224XEU6 A0A195CS46 A0A151J440 A0A2S2PVN4 A0A1I8N3W4 A0A0L0BL65 A0A0V0G9A2 T1PP27 A0A2S2RA84 A0A0Q9XCK0 A0A0Q9XDI8 A0A0P8XU96 A0A3L8DXE8 A0A0Q9WRZ5 A0A0Q9WT90 A0A0Q9X344 A0A2H8TMG4 J9JN11 A0A2S2NL54 A0A182IJC7
Pubmed
22118469
26354079
23622113
26227816
24845553
29403074
+ More
24945155 26483478 25244985 12364791 14747013 17210077 17510324 18362917 19820115 20920257 23761445 28004739 23537049 20566863 26369729 21719571 20798317 24508170 21347285 20353571 21292972 23691247 25315136 25474469 27129103 26334808 26108605 17994087 30249741
24945155 26483478 25244985 12364791 14747013 17210077 17510324 18362917 19820115 20920257 23761445 28004739 23537049 20566863 26369729 21719571 20798317 24508170 21347285 20353571 21292972 23691247 25315136 25474469 27129103 26334808 26108605 17994087 30249741
EMBL
AGBW02008828
OWR52454.1
ODYU01004284
SOQ43990.1
KQ459232
KPJ02710.1
+ More
NWSH01002300 PCG68589.1 PCG68588.1 PCG68591.1 GAIX01013910 JAA78650.1 KQ461155 KPJ08223.1 JTDY01000251 KOB78060.1 KK852980 KDR12978.1 GFDL01003766 JAV31279.1 GFDL01012451 JAV22594.1 PYGN01001777 PSN32859.1 GFDF01002838 JAV11246.1 NEVH01003493 PNF40728.1 GANO01002391 JAB57480.1 JXUM01036387 JXUM01076281 GAPW01002261 KQ562936 KQ561093 JAC11337.1 KXJ74777.1 KXJ79725.1 DS233550 DS232206 EDS30501.1 EDS37452.1 AXCN02001284 AXCM01009690 AAAB01008811 EAA04922.4 EGK96275.1 EGK96276.1 APCN01004735 EGK96277.1 CH477387 EAT42039.1 EAT42040.1 KQ971363 EFA07775.1 GEBQ01026290 JAT13687.1 GGFM01007412 MBW28163.1 GGFK01007546 MBW40867.1 GGFJ01006440 MBW55581.1 ADMH02001125 ETN64039.1 GEZM01007559 JAV95091.1 GGFM01004009 MBW24760.1 GGFK01007575 MBW40896.1 GGFK01007576 MBW40897.1 GGFJ01006565 MBW55706.1 GGFK01007574 MBW40895.1 GEDC01008434 JAS28864.1 KQ434869 KZC09362.1 KZ288353 PBC27332.1 KQ435916 KOX68809.1 APGK01058769 KB741291 KB631759 ENN70623.1 ERL85839.1 DS235093 EEB11855.1 GDAI01000254 JAI17349.1 GDIP01008084 JAM95631.1 GL888120 EGI66985.1 GL438128 EFN69532.1 KK107161 EZA56572.1 GBYB01008647 JAG78414.1 KQ976719 KYM76746.1 ADTU01024933 LRGB01001361 KZS12441.1 EZ422280 ADD18556.1 GDIQ01255571 JAJ96153.1 GL732526 EFX88424.1 CCAG010010739 CCAG010010740 CP012526 ALC47615.1 AK417614 BAN20829.1 KA647242 AFP61871.1 JXJN01013194 GBBI01000555 JAC18157.1 GDKW01000454 JAI56141.1 GBGD01001918 JAC86971.1 ACPB03014184 GAHY01000518 JAA76992.1 GFTR01005429 JAW10997.1 KQ977329 KYN03558.1 KQ980211 KYN17206.1 GGMS01000201 MBY69404.1 JRES01001702 KNC20806.1 GECL01002114 JAP04010.1 KA649748 AFP64377.1 GGMS01017059 MBY86262.1 CH933806 KRG02238.1 KRG02247.1 CH902623 KPU72887.1 QOIP01000003 RLU25121.1 CH940650 KRF83355.1 KRF83360.1 KRG02245.1 GFXV01003510 MBW15315.1 ABLF02028715 GGMR01005286 MBY17905.1
NWSH01002300 PCG68589.1 PCG68588.1 PCG68591.1 GAIX01013910 JAA78650.1 KQ461155 KPJ08223.1 JTDY01000251 KOB78060.1 KK852980 KDR12978.1 GFDL01003766 JAV31279.1 GFDL01012451 JAV22594.1 PYGN01001777 PSN32859.1 GFDF01002838 JAV11246.1 NEVH01003493 PNF40728.1 GANO01002391 JAB57480.1 JXUM01036387 JXUM01076281 GAPW01002261 KQ562936 KQ561093 JAC11337.1 KXJ74777.1 KXJ79725.1 DS233550 DS232206 EDS30501.1 EDS37452.1 AXCN02001284 AXCM01009690 AAAB01008811 EAA04922.4 EGK96275.1 EGK96276.1 APCN01004735 EGK96277.1 CH477387 EAT42039.1 EAT42040.1 KQ971363 EFA07775.1 GEBQ01026290 JAT13687.1 GGFM01007412 MBW28163.1 GGFK01007546 MBW40867.1 GGFJ01006440 MBW55581.1 ADMH02001125 ETN64039.1 GEZM01007559 JAV95091.1 GGFM01004009 MBW24760.1 GGFK01007575 MBW40896.1 GGFK01007576 MBW40897.1 GGFJ01006565 MBW55706.1 GGFK01007574 MBW40895.1 GEDC01008434 JAS28864.1 KQ434869 KZC09362.1 KZ288353 PBC27332.1 KQ435916 KOX68809.1 APGK01058769 KB741291 KB631759 ENN70623.1 ERL85839.1 DS235093 EEB11855.1 GDAI01000254 JAI17349.1 GDIP01008084 JAM95631.1 GL888120 EGI66985.1 GL438128 EFN69532.1 KK107161 EZA56572.1 GBYB01008647 JAG78414.1 KQ976719 KYM76746.1 ADTU01024933 LRGB01001361 KZS12441.1 EZ422280 ADD18556.1 GDIQ01255571 JAJ96153.1 GL732526 EFX88424.1 CCAG010010739 CCAG010010740 CP012526 ALC47615.1 AK417614 BAN20829.1 KA647242 AFP61871.1 JXJN01013194 GBBI01000555 JAC18157.1 GDKW01000454 JAI56141.1 GBGD01001918 JAC86971.1 ACPB03014184 GAHY01000518 JAA76992.1 GFTR01005429 JAW10997.1 KQ977329 KYN03558.1 KQ980211 KYN17206.1 GGMS01000201 MBY69404.1 JRES01001702 KNC20806.1 GECL01002114 JAP04010.1 KA649748 AFP64377.1 GGMS01017059 MBY86262.1 CH933806 KRG02238.1 KRG02247.1 CH902623 KPU72887.1 QOIP01000003 RLU25121.1 CH940650 KRF83355.1 KRF83360.1 KRG02245.1 GFXV01003510 MBW15315.1 ABLF02028715 GGMR01005286 MBY17905.1
Proteomes
UP000007151
UP000053268
UP000218220
UP000053240
UP000037510
UP000027135
+ More
UP000245037 UP000235965 UP000069940 UP000249989 UP000002320 UP000192223 UP000075886 UP000076408 UP000075883 UP000075885 UP000075903 UP000076407 UP000007062 UP000075840 UP000008820 UP000007266 UP000075920 UP000000673 UP000069272 UP000075884 UP000076502 UP000242457 UP000053105 UP000005203 UP000019118 UP000030742 UP000009046 UP000007755 UP000095300 UP000000311 UP000053097 UP000078540 UP000005205 UP000076858 UP000000305 UP000091820 UP000092444 UP000092553 UP000092443 UP000095301 UP000078200 UP000092460 UP000092445 UP000015103 UP000078542 UP000078492 UP000037069 UP000009192 UP000007801 UP000279307 UP000008792 UP000007819 UP000075880
UP000245037 UP000235965 UP000069940 UP000249989 UP000002320 UP000192223 UP000075886 UP000076408 UP000075883 UP000075885 UP000075903 UP000076407 UP000007062 UP000075840 UP000008820 UP000007266 UP000075920 UP000000673 UP000069272 UP000075884 UP000076502 UP000242457 UP000053105 UP000005203 UP000019118 UP000030742 UP000009046 UP000007755 UP000095300 UP000000311 UP000053097 UP000078540 UP000005205 UP000076858 UP000000305 UP000091820 UP000092444 UP000092553 UP000092443 UP000095301 UP000078200 UP000092460 UP000092445 UP000015103 UP000078542 UP000078492 UP000037069 UP000009192 UP000007801 UP000279307 UP000008792 UP000007819 UP000075880
Interpro
Gene 3D
ProteinModelPortal
A0A212FFD9
A0A2H1VT34
A0A194QCK1
A0A2A4J9R9
A0A2A4JA90
A0A2A4J9G1
+ More
S4NY76 A0A194QRT0 A0A0L7LRH4 A0A067QSW9 A0A1Q3FUK3 A0A1Q3F4W5 A0A2P8XLH9 A0A1L8DXV3 A0A2J7RIR1 U5EJ30 A0A023EPD2 B0WZD8 A0A1W4XDU5 A0A182Q031 A0A182Y555 A0A182M4D5 A0A182P292 A0A182VFU8 A0A182XGN9 F5HJ15 A0A182IDM3 F5HJ16 Q176H6 D6WV67 A0A1B6KQH7 A0A2M3ZI16 A0A182W461 A0A2M4AJF0 A0A2M4BRN0 W5JMP8 A0A1Y1NB29 A0A2M3Z8E2 A0A2M4AJD8 A0A2M4AJH2 A0A182FMC3 A0A2M4BRK1 A0A2M4AJD3 A0A182MZK4 A0A1B6DT63 A0A154PBZ1 A0A2A3E7S4 A0A0M8ZST7 A0A088A2Q4 N6SZF4 E0VEP9 A0A0K8TSJ4 A0A0P6BZV0 F4WFK8 A0A1I8PA62 E2AAI5 A0A026WKJ1 A0A0C9R767 A0A151HYR2 A0A158NSI1 A0A164VLK5 D3TLH8 A0A0N8API6 E9FWJ1 A0A1A9X421 A0A1B0FRE4 A0A0M4EPC1 A0A1A9XT98 R4WDK8 T1PEN3 A0A1A9V4X9 A0A1B0BEZ9 A0A1A9ZZN5 A0A1I8PA56 A0A023F9K4 A0A0P4W463 A0A1I8PA55 A0A069DTA1 R4FNN1 A0A1I8PAA7 A0A224XEU6 A0A195CS46 A0A151J440 A0A2S2PVN4 A0A1I8N3W4 A0A0L0BL65 A0A0V0G9A2 T1PP27 A0A2S2RA84 A0A0Q9XCK0 A0A0Q9XDI8 A0A0P8XU96 A0A3L8DXE8 A0A0Q9WRZ5 A0A0Q9WT90 A0A0Q9X344 A0A2H8TMG4 J9JN11 A0A2S2NL54 A0A182IJC7
S4NY76 A0A194QRT0 A0A0L7LRH4 A0A067QSW9 A0A1Q3FUK3 A0A1Q3F4W5 A0A2P8XLH9 A0A1L8DXV3 A0A2J7RIR1 U5EJ30 A0A023EPD2 B0WZD8 A0A1W4XDU5 A0A182Q031 A0A182Y555 A0A182M4D5 A0A182P292 A0A182VFU8 A0A182XGN9 F5HJ15 A0A182IDM3 F5HJ16 Q176H6 D6WV67 A0A1B6KQH7 A0A2M3ZI16 A0A182W461 A0A2M4AJF0 A0A2M4BRN0 W5JMP8 A0A1Y1NB29 A0A2M3Z8E2 A0A2M4AJD8 A0A2M4AJH2 A0A182FMC3 A0A2M4BRK1 A0A2M4AJD3 A0A182MZK4 A0A1B6DT63 A0A154PBZ1 A0A2A3E7S4 A0A0M8ZST7 A0A088A2Q4 N6SZF4 E0VEP9 A0A0K8TSJ4 A0A0P6BZV0 F4WFK8 A0A1I8PA62 E2AAI5 A0A026WKJ1 A0A0C9R767 A0A151HYR2 A0A158NSI1 A0A164VLK5 D3TLH8 A0A0N8API6 E9FWJ1 A0A1A9X421 A0A1B0FRE4 A0A0M4EPC1 A0A1A9XT98 R4WDK8 T1PEN3 A0A1A9V4X9 A0A1B0BEZ9 A0A1A9ZZN5 A0A1I8PA56 A0A023F9K4 A0A0P4W463 A0A1I8PA55 A0A069DTA1 R4FNN1 A0A1I8PAA7 A0A224XEU6 A0A195CS46 A0A151J440 A0A2S2PVN4 A0A1I8N3W4 A0A0L0BL65 A0A0V0G9A2 T1PP27 A0A2S2RA84 A0A0Q9XCK0 A0A0Q9XDI8 A0A0P8XU96 A0A3L8DXE8 A0A0Q9WRZ5 A0A0Q9WT90 A0A0Q9X344 A0A2H8TMG4 J9JN11 A0A2S2NL54 A0A182IJC7
PDB
2JI4
E-value=1.11305e-132,
Score=1212
Ontologies
GO
PANTHER
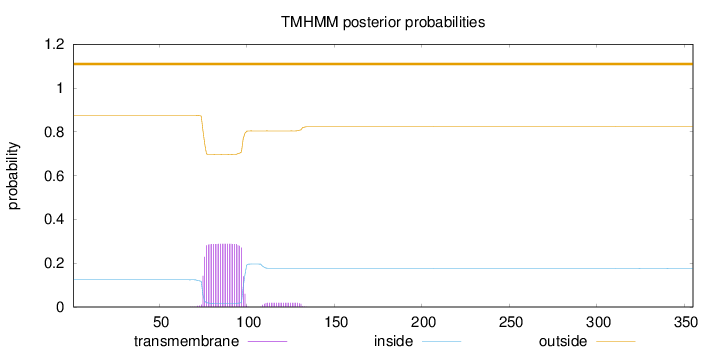
Topology
Length:
355
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.02801
Exp number, first 60 AAs:
0.000780000000000001
Total prob of N-in:
0.12427
outside
1 - 355
Population Genetic Test Statistics
Pi
186.579527
Theta
168.963138
Tajima's D
0.334652
CLR
0.01663
CSRT
0.466626668666567
Interpretation
Uncertain
