Gene
KWMTBOMO10217
Pre Gene Modal
BGIBMGA005599
Annotation
PREDICTED:_tsukushin-like_[Amyelois_transitella]
Full name
Chloride channel protein
Location in the cell
PlasmaMembrane Reliability : 1.737
Sequence
CDS
ATGTATTTTCCGGCTGTGTTAACGATAACAATCGTCCTTGTATCATCAAACCGAGCTGGGGCTGACGCGCCTAGTGCCCCTTGCGATATATCCGTAGAAGATACAAGTAACCTTACGCTGAACTGCTCACGACGAGACATCGTTAAGATACCTAATTGGCCGGAACAAATAAACAACATAAGCAAAGATGGTCACGTCCTAGTGACGTTTTTCAACAACAGCATTACAAATATAACGCAACTGAGTCCGATACCGGGGAACAGGATCGCGATCAGTTTCAAATCAAATCGCGTTATTGAAATCGAAGATGACGCTTTCAGGAACATCCCGCGATTGGTCTATTTGGATCTCAGCAATAATAAAATTTCAGGTCAAGTTCTCAGGAGTGAGATATTCCAAGGACCTTACGAAGATGGGATCTACCGTGAGATTGCTTTGGAGACACTCAATCTTGGTTATAATGAGATCCACTCATTGGACAGATATTTGTTTAAATATACGCCGAACTTAACGAGATTGTACTTAAACAATAACCCTATTGAAATACTTGACCACGTTTCAATACTCGCCATTTCTAGCGCCACGAATCTAGAGGTTCTCGACTTGTCCAAAACCGATATCGACAGTATACCATTGGACGCTTTTAGAGGATTGGTAAAATTACAACAGATCGATTTATCGGATAATCATTTTGTAACCGTACCTGAGAGTTTGAGCCTCGTAGGGAGCACCCTCAAATACCTCACGTTCAACAATAATCCCGTTGTGGAACTGAACGACGACAGTTTCGTAGGATTGTCAAATGTGATAGAATTAGAGGTAGGCGAAAACGATTATTTGGACGAAGTTAAACGGGGCACATTCACACCTTTGAAAAGCTTGAAGGTCCTGCATTTATGTCACAATCGCAACTTGCGATATATCAGTCACAATGCTTTTAGGGGACTAAAAGATAAGTGGACCTTGAAGGAGGTGTATCTAGATGACAATAATTTAAGTGAGTTGTCTCCTGATCTTATGCCATGGAGCAAACTGGATGTTCTCGGGATGACTGGTAATAGCTGGCTGTGTAATTGTGATCTGGTCAACATTGTTAATAAACAGGGTGCTGGTGATAAGTTCAAGTCTGATGAAGTCCCTTATTGTGCTGCTCCTATGAGATTGAGTGGAGAATACATTACAAATATTACTATCAACTTTTGTCCAATGTTAGACAAGACATTTTCACCTAGGAAGACTTTTAGTTTATCCGATTTGAGGCCAAAACATGTTCTATGGACAATCCTTGCCGTTGCAATGGTAGCTGCGTTTGGCATGATCTTGGGACTTCTTGTTAATGCAATCAAGAAAGTATATAAGACAAGGTTCCAAAGTCAACCAATTCACTATATTAATATTAACTCAGACAGCTCGTTTGCATGA
Protein
MYFPAVLTITIVLVSSNRAGADAPSAPCDISVEDTSNLTLNCSRRDIVKIPNWPEQINNISKDGHVLVTFFNNSITNITQLSPIPGNRIAISFKSNRVIEIEDDAFRNIPRLVYLDLSNNKISGQVLRSEIFQGPYEDGIYREIALETLNLGYNEIHSLDRYLFKYTPNLTRLYLNNNPIEILDHVSILAISSATNLEVLDLSKTDIDSIPLDAFRGLVKLQQIDLSDNHFVTVPESLSLVGSTLKYLTFNNNPVVELNDDSFVGLSNVIELEVGENDYLDEVKRGTFTPLKSLKVLHLCHNRNLRYISHNAFRGLKDKWTLKEVYLDDNNLSELSPDLMPWSKLDVLGMTGNSWLCNCDLVNIVNKQGAGDKFKSDEVPYCAAPMRLSGEYITNITINFCPMLDKTFSPRKTFSLSDLRPKHVLWTILAVAMVAAFGMILGLLVNAIKKVYKTRFQSQPIHYININSDSSFA
Summary
Similarity
Belongs to the chloride channel (TC 2.A.49) family.
Feature
chain Chloride channel protein
Uniprot
A0A2A4JT20
A0A2H1WXZ8
A0A194QCJ6
A0A1E1W5M6
E3UKL3
A0A212FFF3
+ More
A0A0L7LAF9 A0A1I8MDK9 A0A1L8D9W0 A0A1I8PC16 A0A1I8PC88 A0A1I8PC18 A0A1I8PC72 A0A1A9W9I4 A0A2J7Q157 W8APT0 A0A1L8EB73 U5ER79 A0A2J7Q168 A0A1B0GHB3 B4N9V7 A0A0K8VMV8 A0A0K8V9D2 A0A1Y1LS93 A0A0K8W4B8 A0A0L0BKJ2 T1PKF9 A0A0M3QXV9 A0A139WD84 A0A034V8X0 E9GRR5 A0A1B0D0U6 B4JEZ3 A0A1A9ZSS1 A0A1A9UTG6 A0A1A9YDQ1 A0A1B0G197 A0A1B0B320 B0WPC7 A0A1J1HLB3 N6T3Z7 B4K8L5 B4M3X1 A0A0P5B397 V5I8Y4 A0A2S2PYY5 A0A0P4Y3N7 A0A0P5GCU8 A0A0P5BH07 A0A0P5VDS4 A0A0P5A7N5 A0A0P5ZRW5 A0A0P5ZA07 A0A0P5P320 A0A0P5NK76 A0A0P5Y2P3 A0A0P5YUN6 A0A0P6AWK8 A0A336MK02 A0A0P5QN88 A0A0N8CIR4 A0A0P5C7P0 A0A1Q3FHH6 B3NZ34 A0A1W4U840 E0W140 A0A1S4F532 A0A1S4F556 Q17F19 B4I265 Q17F18 B4QU01 A0A0R1E546 B4PM01 Q299N9 A0A0P5QP51 A0A3B0KRV3 B4G5H4 A0A0P5W0V3 A0A0P5X2B6 A0A0P6HAR4 A0A182GWZ3 A0A023EWE0 A0A1D2MFR3 Q9VE49 B3LW74 A0A2S2P2H8 A0A2J7Q0W9
A0A0L7LAF9 A0A1I8MDK9 A0A1L8D9W0 A0A1I8PC16 A0A1I8PC88 A0A1I8PC18 A0A1I8PC72 A0A1A9W9I4 A0A2J7Q157 W8APT0 A0A1L8EB73 U5ER79 A0A2J7Q168 A0A1B0GHB3 B4N9V7 A0A0K8VMV8 A0A0K8V9D2 A0A1Y1LS93 A0A0K8W4B8 A0A0L0BKJ2 T1PKF9 A0A0M3QXV9 A0A139WD84 A0A034V8X0 E9GRR5 A0A1B0D0U6 B4JEZ3 A0A1A9ZSS1 A0A1A9UTG6 A0A1A9YDQ1 A0A1B0G197 A0A1B0B320 B0WPC7 A0A1J1HLB3 N6T3Z7 B4K8L5 B4M3X1 A0A0P5B397 V5I8Y4 A0A2S2PYY5 A0A0P4Y3N7 A0A0P5GCU8 A0A0P5BH07 A0A0P5VDS4 A0A0P5A7N5 A0A0P5ZRW5 A0A0P5ZA07 A0A0P5P320 A0A0P5NK76 A0A0P5Y2P3 A0A0P5YUN6 A0A0P6AWK8 A0A336MK02 A0A0P5QN88 A0A0N8CIR4 A0A0P5C7P0 A0A1Q3FHH6 B3NZ34 A0A1W4U840 E0W140 A0A1S4F532 A0A1S4F556 Q17F19 B4I265 Q17F18 B4QU01 A0A0R1E546 B4PM01 Q299N9 A0A0P5QP51 A0A3B0KRV3 B4G5H4 A0A0P5W0V3 A0A0P5X2B6 A0A0P6HAR4 A0A182GWZ3 A0A023EWE0 A0A1D2MFR3 Q9VE49 B3LW74 A0A2S2P2H8 A0A2J7Q0W9
Pubmed
EMBL
NWSH01000679
PCG74838.1
ODYU01011923
SOQ57939.1
KQ459232
KPJ02705.1
+ More
GDQN01008883 JAT82171.1 HM449887 ADO33022.1 AGBW02008828 OWR52457.1 JTDY01001986 KOB72395.1 GFDF01010826 JAV03258.1 NEVH01019960 PNF22315.1 GAMC01015755 GAMC01015754 JAB90800.1 GFDG01002944 JAV15855.1 GANO01003862 JAB56009.1 PNF22316.1 AJWK01004381 CH964232 EDW81712.1 GDHF01012121 GDHF01008239 JAI40193.1 JAI44075.1 GDHF01016921 GDHF01014523 JAI35393.1 JAI37791.1 GEZM01052547 JAV74446.1 GDHF01006410 JAI45904.1 JRES01001712 KNC20572.1 KA648565 AFP63194.1 CP012526 ALC46603.1 KQ971361 KYB25919.1 GAKP01020945 GAKP01020944 JAC38008.1 GL732560 EFX77868.1 AJVK01021673 AJVK01021674 CH916369 EDV93274.1 CCAG010001934 JXJN01007758 JXJN01007759 DS232022 EDS32302.1 CVRI01000006 CRK88204.1 APGK01044593 APGK01044594 APGK01044595 KB741027 KB632186 ENN74884.1 ERL89729.1 CH933806 EDW14414.1 CH940652 EDW59332.1 GDIP01194935 GDIQ01045617 JAJ28467.1 JAN49120.1 GALX01003880 JAB64586.1 GGMS01001552 MBY70755.1 GDIP01233144 JAI90257.1 GDIQ01242838 JAK08887.1 GDIP01199731 JAJ23671.1 GDIP01101283 JAM02432.1 GDIP01215274 JAJ08128.1 GDIP01225036 GDIP01101284 GDIP01053056 LRGB01003389 JAM50659.1 KZS02855.1 GDIP01047741 JAM55974.1 GDIQ01140784 JAL10942.1 GDIQ01148281 JAL03445.1 GDIP01064811 JAM38904.1 GDIP01054465 JAM49250.1 GDIP01023315 JAM80400.1 UFQS01001369 UFQT01001369 SSX10539.1 SSX30225.1 GDIQ01121782 JAL29944.1 GDIP01127735 JAL75979.1 GDIP01179237 JAJ44165.1 GFDL01008024 JAV27021.1 CH954181 EDV48576.1 DS235866 EEB19346.1 CH477277 EAT45114.1 CH480820 EDW54622.1 EAT45113.1 CM000364 EDX12424.1 CM000160 KRK02802.1 EDW95933.2 CM000070 EAL27664.2 KRT00216.1 KRT00217.1 GDIQ01118222 JAL33504.1 OUUW01000013 SPP87971.1 CH479179 EDW24840.1 GDIP01106889 JAL96825.1 GDIP01090432 JAM13283.1 GDIQ01022319 JAN72418.1 JXUM01094258 KQ564116 KXJ72754.1 GAPW01000854 JAC12744.1 LJIJ01001431 ODM91751.1 AE014297 AY052048 AAF55580.1 AAK93472.1 CH902617 EDV42652.1 GGMR01011040 MBY23659.1 PNF22240.1
GDQN01008883 JAT82171.1 HM449887 ADO33022.1 AGBW02008828 OWR52457.1 JTDY01001986 KOB72395.1 GFDF01010826 JAV03258.1 NEVH01019960 PNF22315.1 GAMC01015755 GAMC01015754 JAB90800.1 GFDG01002944 JAV15855.1 GANO01003862 JAB56009.1 PNF22316.1 AJWK01004381 CH964232 EDW81712.1 GDHF01012121 GDHF01008239 JAI40193.1 JAI44075.1 GDHF01016921 GDHF01014523 JAI35393.1 JAI37791.1 GEZM01052547 JAV74446.1 GDHF01006410 JAI45904.1 JRES01001712 KNC20572.1 KA648565 AFP63194.1 CP012526 ALC46603.1 KQ971361 KYB25919.1 GAKP01020945 GAKP01020944 JAC38008.1 GL732560 EFX77868.1 AJVK01021673 AJVK01021674 CH916369 EDV93274.1 CCAG010001934 JXJN01007758 JXJN01007759 DS232022 EDS32302.1 CVRI01000006 CRK88204.1 APGK01044593 APGK01044594 APGK01044595 KB741027 KB632186 ENN74884.1 ERL89729.1 CH933806 EDW14414.1 CH940652 EDW59332.1 GDIP01194935 GDIQ01045617 JAJ28467.1 JAN49120.1 GALX01003880 JAB64586.1 GGMS01001552 MBY70755.1 GDIP01233144 JAI90257.1 GDIQ01242838 JAK08887.1 GDIP01199731 JAJ23671.1 GDIP01101283 JAM02432.1 GDIP01215274 JAJ08128.1 GDIP01225036 GDIP01101284 GDIP01053056 LRGB01003389 JAM50659.1 KZS02855.1 GDIP01047741 JAM55974.1 GDIQ01140784 JAL10942.1 GDIQ01148281 JAL03445.1 GDIP01064811 JAM38904.1 GDIP01054465 JAM49250.1 GDIP01023315 JAM80400.1 UFQS01001369 UFQT01001369 SSX10539.1 SSX30225.1 GDIQ01121782 JAL29944.1 GDIP01127735 JAL75979.1 GDIP01179237 JAJ44165.1 GFDL01008024 JAV27021.1 CH954181 EDV48576.1 DS235866 EEB19346.1 CH477277 EAT45114.1 CH480820 EDW54622.1 EAT45113.1 CM000364 EDX12424.1 CM000160 KRK02802.1 EDW95933.2 CM000070 EAL27664.2 KRT00216.1 KRT00217.1 GDIQ01118222 JAL33504.1 OUUW01000013 SPP87971.1 CH479179 EDW24840.1 GDIP01106889 JAL96825.1 GDIP01090432 JAM13283.1 GDIQ01022319 JAN72418.1 JXUM01094258 KQ564116 KXJ72754.1 GAPW01000854 JAC12744.1 LJIJ01001431 ODM91751.1 AE014297 AY052048 AAF55580.1 AAK93472.1 CH902617 EDV42652.1 GGMR01011040 MBY23659.1 PNF22240.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000037510
UP000095301
UP000095300
+ More
UP000091820 UP000235965 UP000092461 UP000007798 UP000037069 UP000092553 UP000007266 UP000000305 UP000092462 UP000001070 UP000092445 UP000078200 UP000092443 UP000092444 UP000092460 UP000002320 UP000183832 UP000019118 UP000030742 UP000009192 UP000008792 UP000076858 UP000008711 UP000192221 UP000009046 UP000008820 UP000001292 UP000000304 UP000002282 UP000001819 UP000268350 UP000008744 UP000069940 UP000249989 UP000094527 UP000000803 UP000007801
UP000091820 UP000235965 UP000092461 UP000007798 UP000037069 UP000092553 UP000007266 UP000000305 UP000092462 UP000001070 UP000092445 UP000078200 UP000092443 UP000092444 UP000092460 UP000002320 UP000183832 UP000019118 UP000030742 UP000009192 UP000008792 UP000076858 UP000008711 UP000192221 UP000009046 UP000008820 UP000001292 UP000000304 UP000002282 UP000001819 UP000268350 UP000008744 UP000069940 UP000249989 UP000094527 UP000000803 UP000007801
PRIDE
Pfam
Interpro
IPR032675
LRR_dom_sf
+ More
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR026906 LRR_5
IPR001807 Cl-channel_volt-gated
IPR014743 Cl-channel_core
IPR000644 CBS_dom
IPR025875 Leu-rich_rpt_4
IPR001020 PTS_HPr_His_P_site
IPR000483 Cys-rich_flank_reg_C
IPR035427 Tim10-like_dom_sf
IPR004217 Tim10-like
IPR001611 Leu-rich_rpt
IPR003591 Leu-rich_rpt_typical-subtyp
IPR026906 LRR_5
IPR001807 Cl-channel_volt-gated
IPR014743 Cl-channel_core
IPR000644 CBS_dom
IPR025875 Leu-rich_rpt_4
IPR001020 PTS_HPr_His_P_site
IPR000483 Cys-rich_flank_reg_C
IPR035427 Tim10-like_dom_sf
IPR004217 Tim10-like
Gene 3D
ProteinModelPortal
A0A2A4JT20
A0A2H1WXZ8
A0A194QCJ6
A0A1E1W5M6
E3UKL3
A0A212FFF3
+ More
A0A0L7LAF9 A0A1I8MDK9 A0A1L8D9W0 A0A1I8PC16 A0A1I8PC88 A0A1I8PC18 A0A1I8PC72 A0A1A9W9I4 A0A2J7Q157 W8APT0 A0A1L8EB73 U5ER79 A0A2J7Q168 A0A1B0GHB3 B4N9V7 A0A0K8VMV8 A0A0K8V9D2 A0A1Y1LS93 A0A0K8W4B8 A0A0L0BKJ2 T1PKF9 A0A0M3QXV9 A0A139WD84 A0A034V8X0 E9GRR5 A0A1B0D0U6 B4JEZ3 A0A1A9ZSS1 A0A1A9UTG6 A0A1A9YDQ1 A0A1B0G197 A0A1B0B320 B0WPC7 A0A1J1HLB3 N6T3Z7 B4K8L5 B4M3X1 A0A0P5B397 V5I8Y4 A0A2S2PYY5 A0A0P4Y3N7 A0A0P5GCU8 A0A0P5BH07 A0A0P5VDS4 A0A0P5A7N5 A0A0P5ZRW5 A0A0P5ZA07 A0A0P5P320 A0A0P5NK76 A0A0P5Y2P3 A0A0P5YUN6 A0A0P6AWK8 A0A336MK02 A0A0P5QN88 A0A0N8CIR4 A0A0P5C7P0 A0A1Q3FHH6 B3NZ34 A0A1W4U840 E0W140 A0A1S4F532 A0A1S4F556 Q17F19 B4I265 Q17F18 B4QU01 A0A0R1E546 B4PM01 Q299N9 A0A0P5QP51 A0A3B0KRV3 B4G5H4 A0A0P5W0V3 A0A0P5X2B6 A0A0P6HAR4 A0A182GWZ3 A0A023EWE0 A0A1D2MFR3 Q9VE49 B3LW74 A0A2S2P2H8 A0A2J7Q0W9
A0A0L7LAF9 A0A1I8MDK9 A0A1L8D9W0 A0A1I8PC16 A0A1I8PC88 A0A1I8PC18 A0A1I8PC72 A0A1A9W9I4 A0A2J7Q157 W8APT0 A0A1L8EB73 U5ER79 A0A2J7Q168 A0A1B0GHB3 B4N9V7 A0A0K8VMV8 A0A0K8V9D2 A0A1Y1LS93 A0A0K8W4B8 A0A0L0BKJ2 T1PKF9 A0A0M3QXV9 A0A139WD84 A0A034V8X0 E9GRR5 A0A1B0D0U6 B4JEZ3 A0A1A9ZSS1 A0A1A9UTG6 A0A1A9YDQ1 A0A1B0G197 A0A1B0B320 B0WPC7 A0A1J1HLB3 N6T3Z7 B4K8L5 B4M3X1 A0A0P5B397 V5I8Y4 A0A2S2PYY5 A0A0P4Y3N7 A0A0P5GCU8 A0A0P5BH07 A0A0P5VDS4 A0A0P5A7N5 A0A0P5ZRW5 A0A0P5ZA07 A0A0P5P320 A0A0P5NK76 A0A0P5Y2P3 A0A0P5YUN6 A0A0P6AWK8 A0A336MK02 A0A0P5QN88 A0A0N8CIR4 A0A0P5C7P0 A0A1Q3FHH6 B3NZ34 A0A1W4U840 E0W140 A0A1S4F532 A0A1S4F556 Q17F19 B4I265 Q17F18 B4QU01 A0A0R1E546 B4PM01 Q299N9 A0A0P5QP51 A0A3B0KRV3 B4G5H4 A0A0P5W0V3 A0A0P5X2B6 A0A0P6HAR4 A0A182GWZ3 A0A023EWE0 A0A1D2MFR3 Q9VE49 B3LW74 A0A2S2P2H8 A0A2J7Q0W9
PDB
5MX1
E-value=2.23232e-17,
Score=219
Ontologies
GO
Topology
Subcellular location
Membrane
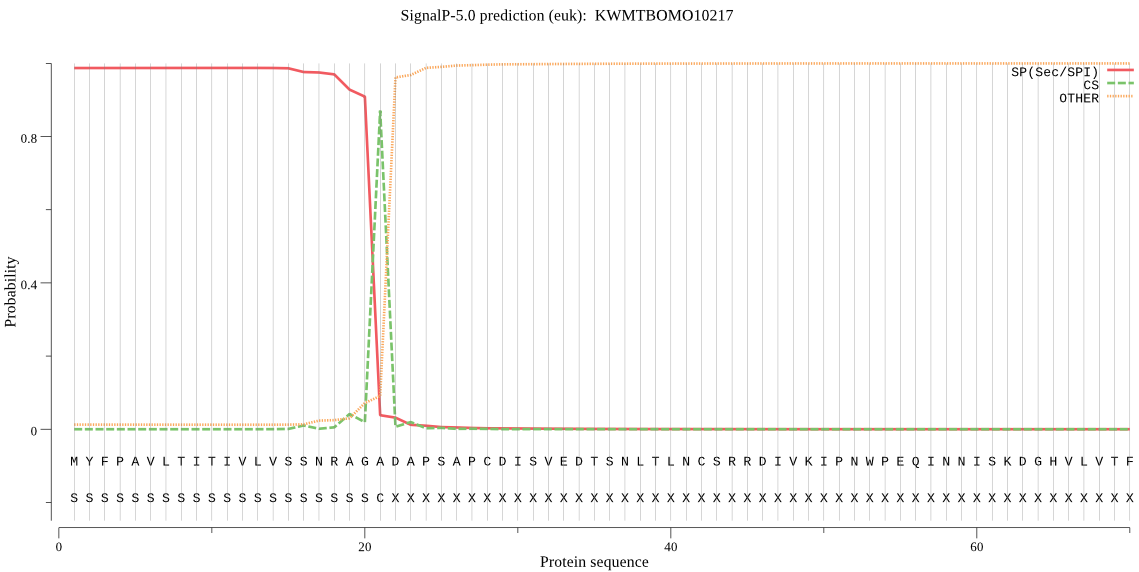
SignalP
Position: 1 - 21,
Likelihood: 0.987003
Length:
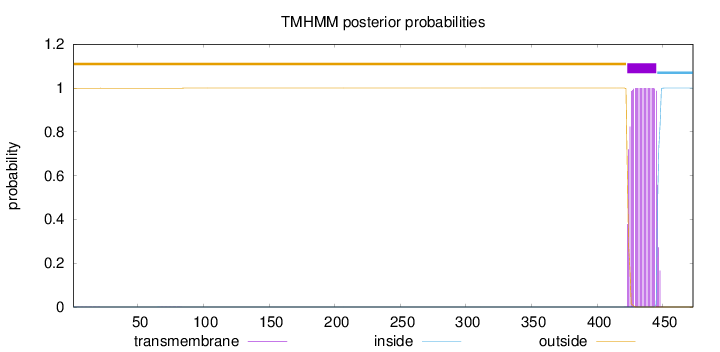
473
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.89777
Exp number, first 60 AAs:
0.01313
Total prob of N-in:
0.00110
outside
1 - 422
TMhelix
423 - 445
inside
446 - 473
Population Genetic Test Statistics
Pi
192.078795
Theta
189.367322
Tajima's D
-1.159841
CLR
1.517767
CSRT
0.110444477776111
Interpretation
Uncertain
