Gene
KWMTBOMO10216
Pre Gene Modal
BGIBMGA005600
Annotation
PREDICTED:_H(+)/Cl(-)_exchange_transporter_7_[Papilio_polytes]
Full name
Chloride channel protein
Location in the cell
PlasmaMembrane Reliability : 4.871
Sequence
CDS
ATGAGTAATAATACTCATTTATCCGATACTGATGAAATAATTTCCGAAACAACATCGGAAACTGCCCAACTTGTTGCCAATGAAGAACCAAGTTCCTCAAATTACACAGAATTAAATAAACCCGATCACGAGGATTCTGATGAAAATGAATTACAGGGTAGTCATGGAGTCCGAAGGAGGAATATTGTTCAAAAGATTGAACCAGGCTCATTGAATGTGTTATCAGCTAAATATGAAAGCTTAGATTATGACACTTGTGAAAATCATCTACTCCTAGATGAAGAACGAAAACGTGGATATCCATTTATAGTTTGGAAGGATATATTGAGATGGTTTATAATTTTGTTGATTGGTATTATCACAGCGCTGATAGCATTTTTCATTGATATTTGCATTGAAGAATTCTCAAATATCAAATATAAAGCCCTCAAGAAATCTGTTGATAAATATGTCTTGCAAGACAAACTATACATTCCATATTTATTATGGGTTTTATCGAATATATGTATAGTGTTTATTGGATCAATGCTAGTTGCATATGTTGAGCCAGTGGCAGCAGGAAGTGGCATACCACAAGTAAAATGTTACCTGAACGGAGTTAAAGTACCTAGAGTAGTAAGGATTAAAACCCTTATTGTAAAAGCCGTAGGTGTCATAACAGCAGTTGTTGGTGGTCTTGCTGGAGGAAAGGAAGGTCCAATGATCCACAGCGGTTCGGTTGTGGCGGCGGGAATATCTCAAGGAAAGAGCACGACGTTCAATAAAGACTTCAAGATATTTCAGTATTTCCGCGAGGATCACGAGAAGAGGGATTTCGTTTCCGCTGGAGCGGCGGCTGGGGTTTCCGCTGCTTTTGGAGCCCCTATAGGCGGTGTGTTATTTTCATTAGAAGAAGGCACTAGTTTTTGGAATCAGGCTCTGACATGGCGAACGTTTTTTGGGACGGTCGTATCCACATTTACTTTGAACTGCGCTTTGTCGGCTTACCACGGCCACCCGGGCGAGCTCAGCTATCCAGGTCTTTTAAATCTGGGTAAAATGGAACCATTCCCATTTCAATTTTACGAGTTGCCAGTATTTATGCTCTTCGGTGTGGTCGGAGGCCTTTTGGGGGCCTTGTGGAATTTTCTCAACTACAAATTGGCGGTGTTTAGAATAAGATACATATGCGCCCCGTGGTTGCGGGTTGTGGAGGCGTGTCTGGTGGCTGCGGCGAGCGCGACCTGCGGTTTTCTGATGATGTTCTTACTCGACGACTGTCGTCCCCTCGGGGAGGACCCTACAAACGTACCCTTACAGTTATACTGCGAAGACGGCGAGTACAACGCGCTAGCCGCGATATGGTTCCAAACGCCCGAAGCGTCGGTGCGATCGTTCCTCCACGACCCGATGGGCTCGTACAAGCCCTGGTCTATACTAATGTTCGTCGTATGCTACTTCCTGCTCTCGACGTGGACGTTCGGTCTGGCCGTTTCAAGCGGCTTGTTCATACCGAACCTGCTGACCGGCGCCGCATGGGGAAGATTGCTCGCTATATCCATACAGTACATGTTACCTTCCAAGACTATTAACCCTGCCAAGTACGCGCTTGTCGGCGCGGCCGCTCAACTGGGAGGCGTCGTACGGATGACGATATCATTAACGGTCATTATTATAGAGACAACGGGACAGATATCAAACGCCCTGCCCATCATCATCACGCTGGTCGTGGCCAAATGGATAGGCGATTTCTTCAATGAAGGTATCTACGATATATATATCCAGCTGGCGGGCGTGCCGCTGTTGCCGTGGGAGCCGCCTCCGTTGGCGCACAACGTGTACGCGTCCGAAGTAATGTCGCATCCGGTGTTCACGTTGCGAACGGTCGAGAACGTTGGACACATCGTCGAGATACTTAAGCTGGTTTCCTACAACGGGTTCCCGGTCGTTGACCCGCCCCTCGCTGATGATTCCGAAGTAACGACATACAAACGTCTTCGTGGACTGATACTGCGCTCCCAGTTAATAGTGCTGTTGCAGAATAAAATATACAATGAAAACGCGAACACGACGTGGTCGAATTTCCAGGTGGACATGGATATGTTCAGGAAGGAATATCCACGATTCCCATCTATAGATGAGCTCGAGATATTAGATTGGGAGAAGTCCTGCATGATAGACCTCCGACCGTTCATGAACCCATCGCCGTACACGTTGCCGCATCGCGCCTCCCTGCCGCGGCTCTTCAGGCTCTTCAGGGCCCTCGGGTTGCGCCACTTGCCCATTGTCAACGATGTCAATGAGGTTGTTGGTATAGTCACTCGGAAAGATATCGCTCGGTACCGAGTATGGAGACATCGAGGACATATGGGAATGGAGGAGTTGGTGCTCTCTAGTGAAATTTAA
Protein
MSNNTHLSDTDEIISETTSETAQLVANEEPSSSNYTELNKPDHEDSDENELQGSHGVRRRNIVQKIEPGSLNVLSAKYESLDYDTCENHLLLDEERKRGYPFIVWKDILRWFIILLIGIITALIAFFIDICIEEFSNIKYKALKKSVDKYVLQDKLYIPYLLWVLSNICIVFIGSMLVAYVEPVAAGSGIPQVKCYLNGVKVPRVVRIKTLIVKAVGVITAVVGGLAGGKEGPMIHSGSVVAAGISQGKSTTFNKDFKIFQYFREDHEKRDFVSAGAAAGVSAAFGAPIGGVLFSLEEGTSFWNQALTWRTFFGTVVSTFTLNCALSAYHGHPGELSYPGLLNLGKMEPFPFQFYELPVFMLFGVVGGLLGALWNFLNYKLAVFRIRYICAPWLRVVEACLVAAASATCGFLMMFLLDDCRPLGEDPTNVPLQLYCEDGEYNALAAIWFQTPEASVRSFLHDPMGSYKPWSILMFVVCYFLLSTWTFGLAVSSGLFIPNLLTGAAWGRLLAISIQYMLPSKTINPAKYALVGAAAQLGGVVRMTISLTVIIIETTGQISNALPIIITLVVAKWIGDFFNEGIYDIYIQLAGVPLLPWEPPPLAHNVYASEVMSHPVFTLRTVENVGHIVEILKLVSYNGFPVVDPPLADDSEVTTYKRLRGLILRSQLIVLLQNKIYNENANTTWSNFQVDMDMFRKEYPRFPSIDELEILDWEKSCMIDLRPFMNPSPYTLPHRASLPRLFRLFRALGLRHLPIVNDVNEVVGIVTRKDIARYRVWRHRGHMGMEELVLSSEI
Summary
Similarity
Belongs to the chloride channel (TC 2.A.49) family.
Feature
chain Chloride channel protein
Uniprot
H9J7V8
A0A2H1WXZ8
A0A194QAY4
A0A3S2N8N5
A0A212FFF2
A0A2A4JS19
+ More
A0A194QRT4 N6TJ32 D6WVA0 A0A1W4WIY7 A0A1Y1MTN9 A0A088AJX8 A0A2A3EC17 U4U608 E9IKJ6 E2BSP0 A0A2J7QWJ5 A0A0L7REZ7 F4WUM4 A0A067RPL9 A0A151JX47 A0A151IG54 E2AWU9 A0A0C9QHE3 A0A195DKZ3 A0A151I5X0 A0A158P195 A0A232EMV2 A0A3L8DN80 A0A1D2NJQ2 A0A310SKQ8 A0A1B6CCK6 A0A1B6L5L6 A0A154PIX8 A0A226E2X4 A0A1B6GUY9 A0A224XF66 A0A1B6HWZ7 J9JTB1 A0A0V0GAG6 A0A2H8TD70 A0A0C9R3S7 A0A026W0K6 A0A0P4WDB3 E0VT99 T1HR68 A0A1E1X2H3 A0A1S3H599 A0A1B6JQE8 A0A3S3P969 A0A0A9YXC6 A0A0A9YRE8 A0A0A9YSZ0 A0A1B6MD94 A0A0K8T5F2 A0A218U9B4 A0A3L8SHD3 U3JNL4 A0A336KZX9 U5EVY1 R7TG07 A0A1V4KTG5 A0A151NGR6 A0A3Q0FT54 T1J037 A0A093PM06 A0A091UT94 A0A091QQH9 J9JJU5 L7M0M8 B0WNS6 A0A131YHF8 A0A1Q3FAT1 A0A146VYY8 A0A224YVS4 A0A146VY27 V9K8E4 F6PY30 A0A293LLE6 A0A146ZWN3 V3ZIM1 A0A146VY76 U3IIS6 A0A3Q1EHE8 A0A3Q1BIG7 A0A3Q1CIX7 A0A3B5ATX8 A0A0P7XRC6 A0A1S3EQT4 A0A3P8T1N4 A0A2B4SNM8 A0A023EVT3 A0A3Q1EDY6 A0A0F8CY29 A0A3P8T0Q6 A0A1W5AIM9 A0A3B5A9G8 A0A3Q3G9P2 A0A3Q3G8V0 A0A091XFI0
A0A194QRT4 N6TJ32 D6WVA0 A0A1W4WIY7 A0A1Y1MTN9 A0A088AJX8 A0A2A3EC17 U4U608 E9IKJ6 E2BSP0 A0A2J7QWJ5 A0A0L7REZ7 F4WUM4 A0A067RPL9 A0A151JX47 A0A151IG54 E2AWU9 A0A0C9QHE3 A0A195DKZ3 A0A151I5X0 A0A158P195 A0A232EMV2 A0A3L8DN80 A0A1D2NJQ2 A0A310SKQ8 A0A1B6CCK6 A0A1B6L5L6 A0A154PIX8 A0A226E2X4 A0A1B6GUY9 A0A224XF66 A0A1B6HWZ7 J9JTB1 A0A0V0GAG6 A0A2H8TD70 A0A0C9R3S7 A0A026W0K6 A0A0P4WDB3 E0VT99 T1HR68 A0A1E1X2H3 A0A1S3H599 A0A1B6JQE8 A0A3S3P969 A0A0A9YXC6 A0A0A9YRE8 A0A0A9YSZ0 A0A1B6MD94 A0A0K8T5F2 A0A218U9B4 A0A3L8SHD3 U3JNL4 A0A336KZX9 U5EVY1 R7TG07 A0A1V4KTG5 A0A151NGR6 A0A3Q0FT54 T1J037 A0A093PM06 A0A091UT94 A0A091QQH9 J9JJU5 L7M0M8 B0WNS6 A0A131YHF8 A0A1Q3FAT1 A0A146VYY8 A0A224YVS4 A0A146VY27 V9K8E4 F6PY30 A0A293LLE6 A0A146ZWN3 V3ZIM1 A0A146VY76 U3IIS6 A0A3Q1EHE8 A0A3Q1BIG7 A0A3Q1CIX7 A0A3B5ATX8 A0A0P7XRC6 A0A1S3EQT4 A0A3P8T1N4 A0A2B4SNM8 A0A023EVT3 A0A3Q1EDY6 A0A0F8CY29 A0A3P8T0Q6 A0A1W5AIM9 A0A3B5A9G8 A0A3Q3G9P2 A0A3Q3G8V0 A0A091XFI0
Pubmed
EMBL
BABH01020018
BABH01020019
BABH01020020
ODYU01011923
SOQ57939.1
KQ459232
+ More
KPJ02703.1 RSAL01000200 RVE44465.1 AGBW02008828 OWR52458.1 NWSH01000679 PCG74837.1 KQ461155 KPJ08228.1 APGK01035530 APGK01035531 KB740928 ENN77853.1 KQ971363 EFA07756.1 GEZM01024238 JAV87835.1 KZ288308 PBC28726.1 KB632064 ERL88502.1 GL763991 EFZ18907.1 GL450241 EFN81286.1 NEVH01009765 PNF32956.1 KQ414608 KOC69403.1 GL888373 EGI62105.1 KK852498 KDR22575.1 KQ981619 KYN39241.1 KQ977743 KYN00188.1 GL443425 EFN62069.1 GBYB01000002 GBYB01000019 GBYB01002586 GBYB01003387 JAG69769.1 JAG69786.1 JAG72353.1 JAG73154.1 KQ980762 KYN13555.1 KQ976402 KYM92301.1 ADTU01006144 ADTU01006145 NNAY01003297 OXU19666.1 QOIP01000006 RLU21831.1 LJIJ01000022 ODN05457.1 KQ763581 OAD54955.1 GEDC01026094 JAS11204.1 GEBQ01020955 JAT19022.1 KQ434931 KZC11809.1 LNIX01000007 OXA52085.1 GECZ01003530 JAS66239.1 GFTR01007998 JAW08428.1 GECU01030373 GECU01028523 JAS77333.1 JAS79183.1 ABLF02035743 GECL01000969 JAP05155.1 GFXV01000252 MBW12057.1 GBYB01002585 JAG72352.1 KK107503 EZA49580.1 GDRN01049003 GDRN01049002 JAI66617.1 DS235760 EEB16605.1 ACPB03012182 ACPB03012183 ACPB03012184 GFAC01005760 JAT93428.1 GECU01028872 GECU01006245 JAS78834.1 JAT01462.1 NCKU01006082 RWS03848.1 GBHO01009394 JAG34210.1 GBHO01009393 JAG34211.1 GBHO01009396 JAG34208.1 GEBQ01006123 JAT33854.1 GBRD01005057 JAG60764.1 MUZQ01000587 OWK50228.1 QUSF01000022 RLW01608.1 AGTO01000604 UFQS01001348 UFQT01001348 SSX10456.1 SSX30142.1 GANO01003206 JAB56665.1 AMQN01013183 KB310021 ELT92718.1 LSYS01001700 OPJ87711.1 AKHW03003035 KYO35954.1 JH431731 KL669029 KFW75290.1 KL410017 KFQ93175.1 KK801697 KFQ29234.1 ABLF02035745 GACK01008346 JAA56688.1 DS232014 EDS31917.1 GEDV01009843 JAP78714.1 GFDL01010423 JAV24622.1 GCES01119610 GCES01064106 JAR22217.1 GFPF01006744 MAA17890.1 GCES01064107 JAR22216.1 JW861589 AFO94106.1 GFWV01003969 MAA28699.1 GCES01015665 JAR70658.1 KB203566 ESO84072.1 GCES01064108 JAR22215.1 ADON01130021 ADON01130022 ADON01130023 ADON01130024 ADON01130025 ADON01130026 JARO02000202 KPP79450.1 LSMT01000055 PFX30117.1 GAPW01000285 JAC13313.1 KQ041130 KKF30224.1 KK735055 KFR12021.1
KPJ02703.1 RSAL01000200 RVE44465.1 AGBW02008828 OWR52458.1 NWSH01000679 PCG74837.1 KQ461155 KPJ08228.1 APGK01035530 APGK01035531 KB740928 ENN77853.1 KQ971363 EFA07756.1 GEZM01024238 JAV87835.1 KZ288308 PBC28726.1 KB632064 ERL88502.1 GL763991 EFZ18907.1 GL450241 EFN81286.1 NEVH01009765 PNF32956.1 KQ414608 KOC69403.1 GL888373 EGI62105.1 KK852498 KDR22575.1 KQ981619 KYN39241.1 KQ977743 KYN00188.1 GL443425 EFN62069.1 GBYB01000002 GBYB01000019 GBYB01002586 GBYB01003387 JAG69769.1 JAG69786.1 JAG72353.1 JAG73154.1 KQ980762 KYN13555.1 KQ976402 KYM92301.1 ADTU01006144 ADTU01006145 NNAY01003297 OXU19666.1 QOIP01000006 RLU21831.1 LJIJ01000022 ODN05457.1 KQ763581 OAD54955.1 GEDC01026094 JAS11204.1 GEBQ01020955 JAT19022.1 KQ434931 KZC11809.1 LNIX01000007 OXA52085.1 GECZ01003530 JAS66239.1 GFTR01007998 JAW08428.1 GECU01030373 GECU01028523 JAS77333.1 JAS79183.1 ABLF02035743 GECL01000969 JAP05155.1 GFXV01000252 MBW12057.1 GBYB01002585 JAG72352.1 KK107503 EZA49580.1 GDRN01049003 GDRN01049002 JAI66617.1 DS235760 EEB16605.1 ACPB03012182 ACPB03012183 ACPB03012184 GFAC01005760 JAT93428.1 GECU01028872 GECU01006245 JAS78834.1 JAT01462.1 NCKU01006082 RWS03848.1 GBHO01009394 JAG34210.1 GBHO01009393 JAG34211.1 GBHO01009396 JAG34208.1 GEBQ01006123 JAT33854.1 GBRD01005057 JAG60764.1 MUZQ01000587 OWK50228.1 QUSF01000022 RLW01608.1 AGTO01000604 UFQS01001348 UFQT01001348 SSX10456.1 SSX30142.1 GANO01003206 JAB56665.1 AMQN01013183 KB310021 ELT92718.1 LSYS01001700 OPJ87711.1 AKHW03003035 KYO35954.1 JH431731 KL669029 KFW75290.1 KL410017 KFQ93175.1 KK801697 KFQ29234.1 ABLF02035745 GACK01008346 JAA56688.1 DS232014 EDS31917.1 GEDV01009843 JAP78714.1 GFDL01010423 JAV24622.1 GCES01119610 GCES01064106 JAR22217.1 GFPF01006744 MAA17890.1 GCES01064107 JAR22216.1 JW861589 AFO94106.1 GFWV01003969 MAA28699.1 GCES01015665 JAR70658.1 KB203566 ESO84072.1 GCES01064108 JAR22215.1 ADON01130021 ADON01130022 ADON01130023 ADON01130024 ADON01130025 ADON01130026 JARO02000202 KPP79450.1 LSMT01000055 PFX30117.1 GAPW01000285 JAC13313.1 KQ041130 KKF30224.1 KK735055 KFR12021.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000007151
UP000218220
UP000053240
+ More
UP000019118 UP000007266 UP000192223 UP000005203 UP000242457 UP000030742 UP000008237 UP000235965 UP000053825 UP000007755 UP000027135 UP000078541 UP000078542 UP000000311 UP000078492 UP000078540 UP000005205 UP000215335 UP000279307 UP000094527 UP000076502 UP000198287 UP000007819 UP000053097 UP000009046 UP000015103 UP000085678 UP000285301 UP000197619 UP000276834 UP000016665 UP000014760 UP000190648 UP000050525 UP000189705 UP000053258 UP000053283 UP000002320 UP000002280 UP000030746 UP000016666 UP000257200 UP000257160 UP000261400 UP000034805 UP000081671 UP000265080 UP000225706 UP000192224 UP000261660 UP000053605
UP000019118 UP000007266 UP000192223 UP000005203 UP000242457 UP000030742 UP000008237 UP000235965 UP000053825 UP000007755 UP000027135 UP000078541 UP000078542 UP000000311 UP000078492 UP000078540 UP000005205 UP000215335 UP000279307 UP000094527 UP000076502 UP000198287 UP000007819 UP000053097 UP000009046 UP000015103 UP000085678 UP000285301 UP000197619 UP000276834 UP000016665 UP000014760 UP000190648 UP000050525 UP000189705 UP000053258 UP000053283 UP000002320 UP000002280 UP000030746 UP000016666 UP000257200 UP000257160 UP000261400 UP000034805 UP000081671 UP000265080 UP000225706 UP000192224 UP000261660 UP000053605
Interpro
Gene 3D
ProteinModelPortal
H9J7V8
A0A2H1WXZ8
A0A194QAY4
A0A3S2N8N5
A0A212FFF2
A0A2A4JS19
+ More
A0A194QRT4 N6TJ32 D6WVA0 A0A1W4WIY7 A0A1Y1MTN9 A0A088AJX8 A0A2A3EC17 U4U608 E9IKJ6 E2BSP0 A0A2J7QWJ5 A0A0L7REZ7 F4WUM4 A0A067RPL9 A0A151JX47 A0A151IG54 E2AWU9 A0A0C9QHE3 A0A195DKZ3 A0A151I5X0 A0A158P195 A0A232EMV2 A0A3L8DN80 A0A1D2NJQ2 A0A310SKQ8 A0A1B6CCK6 A0A1B6L5L6 A0A154PIX8 A0A226E2X4 A0A1B6GUY9 A0A224XF66 A0A1B6HWZ7 J9JTB1 A0A0V0GAG6 A0A2H8TD70 A0A0C9R3S7 A0A026W0K6 A0A0P4WDB3 E0VT99 T1HR68 A0A1E1X2H3 A0A1S3H599 A0A1B6JQE8 A0A3S3P969 A0A0A9YXC6 A0A0A9YRE8 A0A0A9YSZ0 A0A1B6MD94 A0A0K8T5F2 A0A218U9B4 A0A3L8SHD3 U3JNL4 A0A336KZX9 U5EVY1 R7TG07 A0A1V4KTG5 A0A151NGR6 A0A3Q0FT54 T1J037 A0A093PM06 A0A091UT94 A0A091QQH9 J9JJU5 L7M0M8 B0WNS6 A0A131YHF8 A0A1Q3FAT1 A0A146VYY8 A0A224YVS4 A0A146VY27 V9K8E4 F6PY30 A0A293LLE6 A0A146ZWN3 V3ZIM1 A0A146VY76 U3IIS6 A0A3Q1EHE8 A0A3Q1BIG7 A0A3Q1CIX7 A0A3B5ATX8 A0A0P7XRC6 A0A1S3EQT4 A0A3P8T1N4 A0A2B4SNM8 A0A023EVT3 A0A3Q1EDY6 A0A0F8CY29 A0A3P8T0Q6 A0A1W5AIM9 A0A3B5A9G8 A0A3Q3G9P2 A0A3Q3G8V0 A0A091XFI0
A0A194QRT4 N6TJ32 D6WVA0 A0A1W4WIY7 A0A1Y1MTN9 A0A088AJX8 A0A2A3EC17 U4U608 E9IKJ6 E2BSP0 A0A2J7QWJ5 A0A0L7REZ7 F4WUM4 A0A067RPL9 A0A151JX47 A0A151IG54 E2AWU9 A0A0C9QHE3 A0A195DKZ3 A0A151I5X0 A0A158P195 A0A232EMV2 A0A3L8DN80 A0A1D2NJQ2 A0A310SKQ8 A0A1B6CCK6 A0A1B6L5L6 A0A154PIX8 A0A226E2X4 A0A1B6GUY9 A0A224XF66 A0A1B6HWZ7 J9JTB1 A0A0V0GAG6 A0A2H8TD70 A0A0C9R3S7 A0A026W0K6 A0A0P4WDB3 E0VT99 T1HR68 A0A1E1X2H3 A0A1S3H599 A0A1B6JQE8 A0A3S3P969 A0A0A9YXC6 A0A0A9YRE8 A0A0A9YSZ0 A0A1B6MD94 A0A0K8T5F2 A0A218U9B4 A0A3L8SHD3 U3JNL4 A0A336KZX9 U5EVY1 R7TG07 A0A1V4KTG5 A0A151NGR6 A0A3Q0FT54 T1J037 A0A093PM06 A0A091UT94 A0A091QQH9 J9JJU5 L7M0M8 B0WNS6 A0A131YHF8 A0A1Q3FAT1 A0A146VYY8 A0A224YVS4 A0A146VY27 V9K8E4 F6PY30 A0A293LLE6 A0A146ZWN3 V3ZIM1 A0A146VY76 U3IIS6 A0A3Q1EHE8 A0A3Q1BIG7 A0A3Q1CIX7 A0A3B5ATX8 A0A0P7XRC6 A0A1S3EQT4 A0A3P8T1N4 A0A2B4SNM8 A0A023EVT3 A0A3Q1EDY6 A0A0F8CY29 A0A3P8T0Q6 A0A1W5AIM9 A0A3B5A9G8 A0A3Q3G9P2 A0A3Q3G8V0 A0A091XFI0
PDB
6QVU
E-value=2.54394e-28,
Score=315
Ontologies
GO
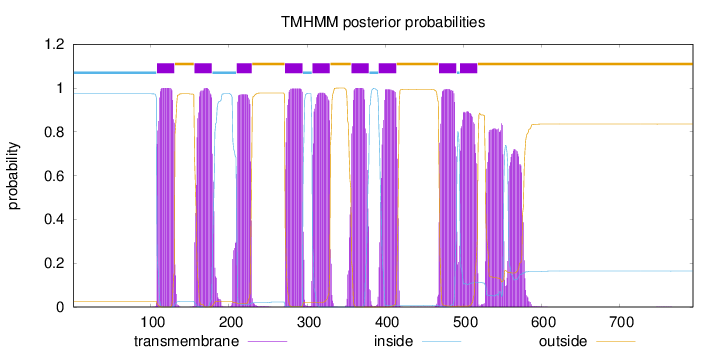
Topology
Subcellular location
Membrane
Length:
794
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
228.2613
Exp number, first 60 AAs:
0
Total prob of N-in:
0.97363
inside
1 - 107
TMhelix
108 - 130
outside
131 - 155
TMhelix
156 - 178
inside
179 - 209
TMhelix
210 - 229
outside
230 - 271
TMhelix
272 - 294
inside
295 - 306
TMhelix
307 - 329
outside
330 - 356
TMhelix
357 - 379
inside
380 - 391
TMhelix
392 - 414
outside
415 - 468
TMhelix
469 - 491
inside
492 - 495
TMhelix
496 - 518
outside
519 - 794
Population Genetic Test Statistics
Pi
220.491035
Theta
165.784205
Tajima's D
1.749031
CLR
0.000175
CSRT
0.837358132093395
Interpretation
Uncertain
