Pre Gene Modal
BGIBMGA005593
Annotation
putative_signal_recognition_particle_68_kDa_protein_[Bombyx_mori]
Full name
Signal recognition particle subunit SRP68
Alternative Name
Signal recognition particle 68 kDa protein
Location in the cell
Cytoplasmic Reliability : 2.347
Sequence
CDS
ATGGTAGGTCAAGAGAGTGAAATTGAAAATGGAAAGGTGACAGCGGTAGAAAATTCTGATAAGAAAAATGAGAAGGCACCCATCCTCCTCAACTTAGAAATATTTCGGATAACAAGAGACAGTCAGCAGCAGCATGGTCTCCGACATGCTGATTATCAGAGATATCGTGGCTATTGTTCGCGCCGCATAAGAAGACTGCGAAAAGTACTCAAAATACCACAGGGAGACAGACGCCATTACCGTCGTCGTGATGTCACAACTACCCACCTTACGGCCAACAATGCCGAGAATAGATTGCTGTACATACCGCTGCTGCAAGCCGAGAGGGCATGGGCTCATGCCATGCAACTGCGTCAGGAAGCTAACACTGAACCGAGAAAAAAGTTTCATCTTGTGTCTCGACTGAAGAAAGCATGTGCACATGGGCAGATGCTTTTGCAGCTTTGTGAGGAAAGCGGCAGGTGCGAAGCGCGTACGGTGCTAGAGGCGGGCGCGTACGCTTCTTGGCTGCGGGGCGTGTTGCTGCTGGAGTTGCAACAATGGCGCGAGGCAGCGGACTCATTGCAACGGGCCAGGATCGTACTCGAACAACTCTGTACAGCCTTGCCGGCGGACGAACGGATTGTTTACGATCAGAAGCTAGAGGAGCTGAAGCCGAGTCTTCGTTATTGTGCTTACAACATCGGTGACGAGTCCGCCGCCGGAGACCTTATGGCCATGCGCGGTCAAGGACTCATGCACAACCTCGATGCGCTGATGGCACAGGCTAAAGAGTCGCGTTCTGGCATTATGCACGAAGTGGAGTGGCGGGGCCGTCGCGTCACTGTCAAACCGGAAAAGGTTCGACTTTTCCTTATTGCTCTACAGGATCTGGACAAATCCGTGTCCAAGGCGGAGACGGCTCAAGCCAAGATCGACATTCTCGAGAACATCCTGATGGACTGCAAGGATGCAATTTCCGCAATCAAGGACGAGATAAAAACCGATCCGAAACTGAAATCCGCACCCGAGATCCAAGCCTCCAGCGTCGGCTACCTGCTCTCGTATTTGATGTACGTGCGACTTGTACGCACTATCGAGCGAAACAATTTGCTCGTCGAACAAGCTGAAGAAGCGCGCAAGAACAACATACAGATCGACGGAAAGAAAGTCCGGCCTCAAGACTTGACCAGGCTCTACGAGATCATTCTGCAGAATTACAATGAACTGCAACAGTTGCCGGGCTTTGAGAACGACGCAGTTTATCAAAAGGAAATCGATACGCAAATGAAGGCGTATCGCGCGTTCCGTTGCTATTACATCGCTCAAGTGCTCACCGGACTGAGACGCTTCAGGGAGGCTTTAGCGATGCTCGAAAGATGCAACACGTATACCGTTGAATCGCTCAAAAGTAAACTGGACAATCAACAACTAAAGGACAAGTTGCAAGTCTTGAAGAAGGACATAGAAAGCTGCAAATTCGAAGTCCACGCTGATTCTGTGCTCGAGGATGATGAAGAAGACGAAGAGAGTAAATACGCTTCGAGCGGCAAATCTTACAAAGACAAGAAGCCTCTGGTGGAGCGCTTGGACGATTACAGAGAAGATTCCCAGGTGCTTACGAAGAACCCGAACATTTATAGAATGCCTCCCCCAATGGAAGCGATTCCTTGTAAGCCACTGTTCTTTGATTTGGCCTGCAATTTCGTTGAGTTCCCGAATTTGGACGATAAAACGGGCGCTACAGATGGCAAGAAGCAAGGTGCAGGACTCACTGGACTCGTTAAAGGATTTTTGGGTTGGGGAAAAAGCGATAAATAG
Protein
MVGQESEIENGKVTAVENSDKKNEKAPILLNLEIFRITRDSQQQHGLRHADYQRYRGYCSRRIRRLRKVLKIPQGDRRHYRRRDVTTTHLTANNAENRLLYIPLLQAERAWAHAMQLRQEANTEPRKKFHLVSRLKKACAHGQMLLQLCEESGRCEARTVLEAGAYASWLRGVLLLELQQWREAADSLQRARIVLEQLCTALPADERIVYDQKLEELKPSLRYCAYNIGDESAAGDLMAMRGQGLMHNLDALMAQAKESRSGIMHEVEWRGRRVTVKPEKVRLFLIALQDLDKSVSKAETAQAKIDILENILMDCKDAISAIKDEIKTDPKLKSAPEIQASSVGYLLSYLMYVRLVRTIERNNLLVEQAEEARKNNIQIDGKKVRPQDLTRLYEIILQNYNELQQLPGFENDAVYQKEIDTQMKAYRAFRCYYIAQVLTGLRRFREALAMLERCNTYTVESLKSKLDNQQLKDKLQVLKKDIESCKFEVHADSVLEDDEEDEESKYASSGKSYKDKKPLVERLDDYREDSQVLTKNPNIYRMPPPMEAIPCKPLFFDLACNFVEFPNLDDKTGATDGKKQGAGLTGLVKGFLGWGKSDK
Summary
Description
Signal-recognition-particle assembly has a crucial role in targeting secretory proteins to the rough endoplasmic reticulum membrane.
Signal-recognition-particle assembly has a crucial role in targeting secretory proteins to the rough endoplasmic reticulum membrane. Srp68 binds the 7S RNA, Srp72 binds to this complex subsequently. This ribonucleoprotein complex might interact directly with the docking protein in the ER membrane and possibly participate in the elongation arrest function (By similarity).
Signal-recognition-particle assembly has a crucial role in targeting secretory proteins to the rough endoplasmic reticulum membrane. Srp68 binds the 7S RNA, Srp72 binds to this complex subsequently. This ribonucleoprotein complex might interact directly with the docking protein in the ER membrane and possibly participate in the elongation arrest function (By similarity).
Subunit
Signal recognition particle consists of a 7S RNA molecule of 300 nucleotides and six protein subunits: Srp72, Srp68, Srp54, Srp19, Srp14 and Srp9.
Similarity
Belongs to the SRP68 family.
Keywords
Complete proteome
Cytoplasm
Reference proteome
Ribonucleoprotein
RNA-binding
Signal recognition particle
Feature
chain Signal recognition particle subunit SRP68
Uniprot
Q0N2R7
H9J7V1
A0A2A4JST9
A0A2H1WXZ3
S4PYH2
A0A212FFF5
+ More
A0A194QCI9 A0A194QRW6 A0A0L7KJZ3 A0A1E1VXV0 A0A1J1IBL8 A0A0K8TT17 A0A1B0GN56 A0A336KGJ9 Q16U06 U5EUQ1 A0A182H0I8 A0A182GPN0 A0A1L8DVJ5 A0A1L8DVZ8 B0WMF9 A0A182Q8R5 A0A1Q3FAB6 A0A2J7PLR9 A0A1B6CIN9 A0A023EU35 A0A1A9UJE0 A0A1A9Z2P3 A0A1B0G0V2 T1PBK6 A0A1A9YEN1 A0A1B0B189 A0A1B0CSX8 D3TL53 A0A0L0CKE1 A0A182XWI8 A0A067RMM2 A0A182PD57 A0A084VXE4 W8BUY5 A0A034VAW1 A0A0K8WL07 A0A182K5W3 A0A182RFV1 A0A182LVI5 A0A1A9WTH5 A0A1I8PHA1 A0A182VYD5 A0A182SJZ4 A0A2M4BHC2 A0A2M4BH93 A0A2M3ZHL6 E0VTC5 W5J3V1 A0A182FA82 A0A2M4BI81 A0A182NMV4 B4N6N3 A0A2M4AAH0 A0A0M3QW49 A0A0A1WUL3 Q7QK73 A0A182HZN0 A0A182WZI3 A0A182LQP2 A0A182V143 A0A182U418 Q29F46 B4HKJ1 A0A3B0JY57 A0A0J9RRS5 A0A1W4V0E4 B4PFG3 A0A1B6L3X3 B4J152 B4KVN8 B3NBK9 B4LEV6 B4QMN5 A0A1S4EAW6 A0A2J7PLS8 Q9VSS2 B3M5W5 A0A182J126 B4HAZ5 A0A026X0Y1 E9IQ46 A0A0P4VSP3 A0A2M3ZHZ3 A0A0N0BED1 A0A1L8DVC2 A0A158P1B8 A0A069DW85 A0A088AR84 E2ATX5 A0A0J7KUK6 E2BQK6 A0A2A3E9E5 J9K2B9 T1I0Y0
A0A194QCI9 A0A194QRW6 A0A0L7KJZ3 A0A1E1VXV0 A0A1J1IBL8 A0A0K8TT17 A0A1B0GN56 A0A336KGJ9 Q16U06 U5EUQ1 A0A182H0I8 A0A182GPN0 A0A1L8DVJ5 A0A1L8DVZ8 B0WMF9 A0A182Q8R5 A0A1Q3FAB6 A0A2J7PLR9 A0A1B6CIN9 A0A023EU35 A0A1A9UJE0 A0A1A9Z2P3 A0A1B0G0V2 T1PBK6 A0A1A9YEN1 A0A1B0B189 A0A1B0CSX8 D3TL53 A0A0L0CKE1 A0A182XWI8 A0A067RMM2 A0A182PD57 A0A084VXE4 W8BUY5 A0A034VAW1 A0A0K8WL07 A0A182K5W3 A0A182RFV1 A0A182LVI5 A0A1A9WTH5 A0A1I8PHA1 A0A182VYD5 A0A182SJZ4 A0A2M4BHC2 A0A2M4BH93 A0A2M3ZHL6 E0VTC5 W5J3V1 A0A182FA82 A0A2M4BI81 A0A182NMV4 B4N6N3 A0A2M4AAH0 A0A0M3QW49 A0A0A1WUL3 Q7QK73 A0A182HZN0 A0A182WZI3 A0A182LQP2 A0A182V143 A0A182U418 Q29F46 B4HKJ1 A0A3B0JY57 A0A0J9RRS5 A0A1W4V0E4 B4PFG3 A0A1B6L3X3 B4J152 B4KVN8 B3NBK9 B4LEV6 B4QMN5 A0A1S4EAW6 A0A2J7PLS8 Q9VSS2 B3M5W5 A0A182J126 B4HAZ5 A0A026X0Y1 E9IQ46 A0A0P4VSP3 A0A2M3ZHZ3 A0A0N0BED1 A0A1L8DVC2 A0A158P1B8 A0A069DW85 A0A088AR84 E2ATX5 A0A0J7KUK6 E2BQK6 A0A2A3E9E5 J9K2B9 T1I0Y0
Pubmed
19121390
23622113
22118469
26354079
26227816
26369729
+ More
17510324 26483478 24945155 25315136 20353571 26108605 25244985 24845553 24438588 24495485 25348373 20566863 20920257 23761445 17994087 25830018 12364791 14747013 17210077 20966253 15632085 22936249 17550304 10731132 12537572 12537569 15901661 24508170 30249741 21282665 27129103 21347285 26334808 20798317
17510324 26483478 24945155 25315136 20353571 26108605 25244985 24845553 24438588 24495485 25348373 20566863 20920257 23761445 17994087 25830018 12364791 14747013 17210077 20966253 15632085 22936249 17550304 10731132 12537572 12537569 15901661 24508170 30249741 21282665 27129103 21347285 26334808 20798317
EMBL
DQ646414
ABH10804.1
BABH01020024
NWSH01000679
PCG74846.1
ODYU01011923
+ More
SOQ57943.1 GAIX01003293 JAA89267.1 AGBW02008828 OWR52462.1 KQ459232 KPJ02700.1 KQ461155 KPJ08232.1 JTDY01009611 KOB63224.1 GDQN01011499 JAT79555.1 CVRI01000045 CRK96956.1 GDAI01000322 JAI17281.1 AJVK01004376 UFQS01000446 UFQT01000446 SSX04036.1 SSX24401.1 CH477635 EAT38012.1 GANO01002223 JAB57648.1 JXUM01101401 KQ564645 KXJ71972.1 JXUM01078749 KQ563085 KXJ74508.1 GFDF01003620 JAV10464.1 GFDF01003619 JAV10465.1 DS231997 EDS31016.1 AXCN02001555 GFDL01010597 JAV24448.1 NEVH01024423 PNF17284.1 GEDC01024018 JAS13280.1 GAPW01000688 JAC12910.1 CCAG010000567 KA646054 AFP60683.1 JXJN01007030 AJWK01026757 EZ422153 ADD18348.1 JRES01000364 KNC31924.1 KK852421 KDR24278.1 ATLV01018026 KE525209 KFB42638.1 GAMC01005777 GAMC01005776 JAC00779.1 GAKP01020025 GAKP01020022 JAC38930.1 GDHF01000744 JAI51570.1 AXCM01004735 GGFJ01003283 MBW52424.1 GGFJ01003284 MBW52425.1 GGFM01007167 MBW27918.1 DS235761 EEB16631.1 ADMH02002183 ETN57993.1 GGFJ01003625 MBW52766.1 CH964161 EDW80022.1 GGFK01004465 MBW37786.1 CP012525 ALC43529.1 GBXI01011740 JAD02552.1 AAAB01008799 EAA03741.4 APCN01001987 CH379068 EAL31226.2 CH480815 EDW40794.1 OUUW01000009 SPP85362.1 CM002912 KMY98526.1 CM000159 EDW93089.1 GEBQ01021636 JAT18341.1 CH916366 EDV96907.1 CH933809 EDW19509.1 CH954178 EDV50605.1 CH940647 EDW70213.1 CM000363 EDX09789.1 PNF17286.1 AE014296 AY058698 CH902618 EDV39655.1 CH479246 EDW37789.1 KK107046 QOIP01000004 EZA61738.1 RLU24201.1 GL764659 EFZ17326.1 GDKW01000430 JAI56165.1 GGFM01007297 MBW28048.1 KQ435837 KOX71472.1 GFDF01003708 JAV10376.1 ADTU01006214 GBGD01000957 JAC87932.1 GL442747 EFN63106.1 LBMM01003073 KMQ93998.1 GL449769 EFN82064.1 KZ288313 PBC28348.1 ABLF02013100 ACPB03010572
SOQ57943.1 GAIX01003293 JAA89267.1 AGBW02008828 OWR52462.1 KQ459232 KPJ02700.1 KQ461155 KPJ08232.1 JTDY01009611 KOB63224.1 GDQN01011499 JAT79555.1 CVRI01000045 CRK96956.1 GDAI01000322 JAI17281.1 AJVK01004376 UFQS01000446 UFQT01000446 SSX04036.1 SSX24401.1 CH477635 EAT38012.1 GANO01002223 JAB57648.1 JXUM01101401 KQ564645 KXJ71972.1 JXUM01078749 KQ563085 KXJ74508.1 GFDF01003620 JAV10464.1 GFDF01003619 JAV10465.1 DS231997 EDS31016.1 AXCN02001555 GFDL01010597 JAV24448.1 NEVH01024423 PNF17284.1 GEDC01024018 JAS13280.1 GAPW01000688 JAC12910.1 CCAG010000567 KA646054 AFP60683.1 JXJN01007030 AJWK01026757 EZ422153 ADD18348.1 JRES01000364 KNC31924.1 KK852421 KDR24278.1 ATLV01018026 KE525209 KFB42638.1 GAMC01005777 GAMC01005776 JAC00779.1 GAKP01020025 GAKP01020022 JAC38930.1 GDHF01000744 JAI51570.1 AXCM01004735 GGFJ01003283 MBW52424.1 GGFJ01003284 MBW52425.1 GGFM01007167 MBW27918.1 DS235761 EEB16631.1 ADMH02002183 ETN57993.1 GGFJ01003625 MBW52766.1 CH964161 EDW80022.1 GGFK01004465 MBW37786.1 CP012525 ALC43529.1 GBXI01011740 JAD02552.1 AAAB01008799 EAA03741.4 APCN01001987 CH379068 EAL31226.2 CH480815 EDW40794.1 OUUW01000009 SPP85362.1 CM002912 KMY98526.1 CM000159 EDW93089.1 GEBQ01021636 JAT18341.1 CH916366 EDV96907.1 CH933809 EDW19509.1 CH954178 EDV50605.1 CH940647 EDW70213.1 CM000363 EDX09789.1 PNF17286.1 AE014296 AY058698 CH902618 EDV39655.1 CH479246 EDW37789.1 KK107046 QOIP01000004 EZA61738.1 RLU24201.1 GL764659 EFZ17326.1 GDKW01000430 JAI56165.1 GGFM01007297 MBW28048.1 KQ435837 KOX71472.1 GFDF01003708 JAV10376.1 ADTU01006214 GBGD01000957 JAC87932.1 GL442747 EFN63106.1 LBMM01003073 KMQ93998.1 GL449769 EFN82064.1 KZ288313 PBC28348.1 ABLF02013100 ACPB03010572
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
+ More
UP000183832 UP000092462 UP000008820 UP000069940 UP000249989 UP000002320 UP000075886 UP000235965 UP000078200 UP000092445 UP000092444 UP000095301 UP000092443 UP000092460 UP000092461 UP000037069 UP000076408 UP000027135 UP000075885 UP000030765 UP000075881 UP000075900 UP000075883 UP000091820 UP000095300 UP000075920 UP000075901 UP000009046 UP000000673 UP000069272 UP000075884 UP000007798 UP000092553 UP000007062 UP000075840 UP000076407 UP000075882 UP000075903 UP000075902 UP000001819 UP000001292 UP000268350 UP000192221 UP000002282 UP000001070 UP000009192 UP000008711 UP000008792 UP000000304 UP000079169 UP000000803 UP000007801 UP000075880 UP000008744 UP000053097 UP000279307 UP000053105 UP000005205 UP000005203 UP000000311 UP000036403 UP000008237 UP000242457 UP000007819 UP000015103
UP000183832 UP000092462 UP000008820 UP000069940 UP000249989 UP000002320 UP000075886 UP000235965 UP000078200 UP000092445 UP000092444 UP000095301 UP000092443 UP000092460 UP000092461 UP000037069 UP000076408 UP000027135 UP000075885 UP000030765 UP000075881 UP000075900 UP000075883 UP000091820 UP000095300 UP000075920 UP000075901 UP000009046 UP000000673 UP000069272 UP000075884 UP000007798 UP000092553 UP000007062 UP000075840 UP000076407 UP000075882 UP000075903 UP000075902 UP000001819 UP000001292 UP000268350 UP000192221 UP000002282 UP000001070 UP000009192 UP000008711 UP000008792 UP000000304 UP000079169 UP000000803 UP000007801 UP000075880 UP000008744 UP000053097 UP000279307 UP000053105 UP000005205 UP000005203 UP000000311 UP000036403 UP000008237 UP000242457 UP000007819 UP000015103
Pfam
PF16969 SRP68
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
Q0N2R7
H9J7V1
A0A2A4JST9
A0A2H1WXZ3
S4PYH2
A0A212FFF5
+ More
A0A194QCI9 A0A194QRW6 A0A0L7KJZ3 A0A1E1VXV0 A0A1J1IBL8 A0A0K8TT17 A0A1B0GN56 A0A336KGJ9 Q16U06 U5EUQ1 A0A182H0I8 A0A182GPN0 A0A1L8DVJ5 A0A1L8DVZ8 B0WMF9 A0A182Q8R5 A0A1Q3FAB6 A0A2J7PLR9 A0A1B6CIN9 A0A023EU35 A0A1A9UJE0 A0A1A9Z2P3 A0A1B0G0V2 T1PBK6 A0A1A9YEN1 A0A1B0B189 A0A1B0CSX8 D3TL53 A0A0L0CKE1 A0A182XWI8 A0A067RMM2 A0A182PD57 A0A084VXE4 W8BUY5 A0A034VAW1 A0A0K8WL07 A0A182K5W3 A0A182RFV1 A0A182LVI5 A0A1A9WTH5 A0A1I8PHA1 A0A182VYD5 A0A182SJZ4 A0A2M4BHC2 A0A2M4BH93 A0A2M3ZHL6 E0VTC5 W5J3V1 A0A182FA82 A0A2M4BI81 A0A182NMV4 B4N6N3 A0A2M4AAH0 A0A0M3QW49 A0A0A1WUL3 Q7QK73 A0A182HZN0 A0A182WZI3 A0A182LQP2 A0A182V143 A0A182U418 Q29F46 B4HKJ1 A0A3B0JY57 A0A0J9RRS5 A0A1W4V0E4 B4PFG3 A0A1B6L3X3 B4J152 B4KVN8 B3NBK9 B4LEV6 B4QMN5 A0A1S4EAW6 A0A2J7PLS8 Q9VSS2 B3M5W5 A0A182J126 B4HAZ5 A0A026X0Y1 E9IQ46 A0A0P4VSP3 A0A2M3ZHZ3 A0A0N0BED1 A0A1L8DVC2 A0A158P1B8 A0A069DW85 A0A088AR84 E2ATX5 A0A0J7KUK6 E2BQK6 A0A2A3E9E5 J9K2B9 T1I0Y0
A0A194QCI9 A0A194QRW6 A0A0L7KJZ3 A0A1E1VXV0 A0A1J1IBL8 A0A0K8TT17 A0A1B0GN56 A0A336KGJ9 Q16U06 U5EUQ1 A0A182H0I8 A0A182GPN0 A0A1L8DVJ5 A0A1L8DVZ8 B0WMF9 A0A182Q8R5 A0A1Q3FAB6 A0A2J7PLR9 A0A1B6CIN9 A0A023EU35 A0A1A9UJE0 A0A1A9Z2P3 A0A1B0G0V2 T1PBK6 A0A1A9YEN1 A0A1B0B189 A0A1B0CSX8 D3TL53 A0A0L0CKE1 A0A182XWI8 A0A067RMM2 A0A182PD57 A0A084VXE4 W8BUY5 A0A034VAW1 A0A0K8WL07 A0A182K5W3 A0A182RFV1 A0A182LVI5 A0A1A9WTH5 A0A1I8PHA1 A0A182VYD5 A0A182SJZ4 A0A2M4BHC2 A0A2M4BH93 A0A2M3ZHL6 E0VTC5 W5J3V1 A0A182FA82 A0A2M4BI81 A0A182NMV4 B4N6N3 A0A2M4AAH0 A0A0M3QW49 A0A0A1WUL3 Q7QK73 A0A182HZN0 A0A182WZI3 A0A182LQP2 A0A182V143 A0A182U418 Q29F46 B4HKJ1 A0A3B0JY57 A0A0J9RRS5 A0A1W4V0E4 B4PFG3 A0A1B6L3X3 B4J152 B4KVN8 B3NBK9 B4LEV6 B4QMN5 A0A1S4EAW6 A0A2J7PLS8 Q9VSS2 B3M5W5 A0A182J126 B4HAZ5 A0A026X0Y1 E9IQ46 A0A0P4VSP3 A0A2M3ZHZ3 A0A0N0BED1 A0A1L8DVC2 A0A158P1B8 A0A069DW85 A0A088AR84 E2ATX5 A0A0J7KUK6 E2BQK6 A0A2A3E9E5 J9K2B9 T1I0Y0
PDB
6FRK
E-value=1.71526e-110,
Score=1023
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Length:
599
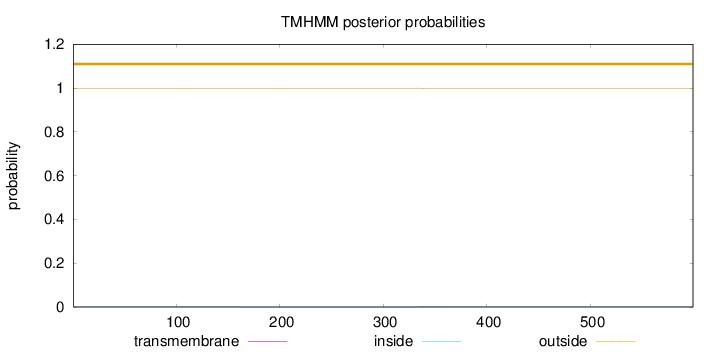
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02261
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00059
outside
1 - 599
Population Genetic Test Statistics
Pi
157.097266
Theta
169.546876
Tajima's D
-0.84594
CLR
0.208874
CSRT
0.165541722913854
Interpretation
Uncertain
