Gene
KWMTBOMO10210
Annotation
PREDICTED:_uncharacterized_protein_LOC106718167_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 1.654
Sequence
CDS
ATGAATGAAAATATATCAGAAATAAAAGGAAGTTCTGACTTTTCCAAACATGATTGCGTCTCCTTTTTGAATCTACTCAACAAATACGTGAAAGTCAAGCTCTTAAAAAATTCAGAATTTGAAGGGTTTATACGGTGTATAGATCCGATATCTCATAGCTTAATTCTTTCCATAGCAAATCACAGCACATACCACACAGTGGTTGTACCCGGCCATGCAATAATAGATGTTACAGAAACACATACAGATTTGATACCACCTATTCAGACACCTGCTAGTTACGTTGAAGACATTAGTGTGGATAGAAAATCTAATCTAATATCTTGGCTGAAAAAGAATTTGTTGCCAGTTACAGAAGTGAATGACAATATTGTATTTGGAAATGTGACGATAATTCCTCCTTATAGTATTAATGATATTTGTACGGATAATACTATTGTGGCAATGCAAATGCGAAAACTAATTCGACAAATGCCATAA
Protein
MNENISEIKGSSDFSKHDCVSFLNLLNKYVKVKLLKNSEFEGFIRCIDPISHSLILSIANHSTYHTVVVPGHAIIDVTETHTDLIPPIQTPASYVEDISVDRKSNLISWLKKNLLPVTEVNDNIVFGNVTIIPPYSINDICTDNTIVAMQMRKLIRQMP
Summary
Uniprot
A0A2A4JTP8
A0A194QTA5
A0A2H1VKA0
A0A194QB57
A0A1E1WFQ1
A0A212F502
+ More
S4PS67 A0A023F8B4 A0A0V0GF60 A0A069DNF8 A0A224XMQ2 A0A0P4VNE1 R4G3X0 A7SXH9 V9LDM0 K4G087 A0A2D0RS50 A0A091R9N1 A0A1A8H3I7 A0A1A8EQS7 A0A1A8MMY7 A0A1A8NDK8 A0A1A8NEX9 A0A1A8S930 A0A3N0XE49 A0A1A8IFV7 A0A1A8JNB7 A0A1A8UKI1 A0A3Q2TVU9 W5KHX1 A0A1A8DJ81 A0A146XFF7 A0A1A7ZC24 A0A1A8CN82 A0A210QSX9 A0A3Q3A060 A0A3B5JYJ6 A0A3S1BB47 R0JWL8 A0A1V4IFV8 A0A3Q2CJI7 H3C699 A0A2R7WCX4 A0A3M6TQY6 A0A163DF35 R7VWK2 V3Z517 A0A3B3D9G8 A0A087V7E0 A0A1A7XF10 A0A087V1A5 A0A131Y815 A0A3B4C3Q8 A0A093CRB9 A0A3Q3GKT4 A0A044S4L3 A0A3P8X1E4 G1N663 A0A3Q1G8G5 A0A093NAX7 A0A087QTH5 A0A2U9BM96 A0A226NLT4 G3WT92 A0A091LHX4 A0A091IMZ1 A0A091P8D4 A0A091R091 A0A2P4SL83 A0A091SW12 A0A3B3U7C6 A0A091NKN4 G1KF12 G3MT35 A0A023GA07 A0A093BK81 W5NFL4 A0A091U9H8 A0A093KAW7 A0A226P6L2 K7FA78 A0A093II94 A0A023G795 E1BRN1 A0A293LAH9 A0A091FUX2 A0A0N4T061 A0A2I0U7D6 A0A0K0ILX7 A0A0R3R313 A0A093J7A2 A0A1S2ZL65 A0A093FY15 A0A224YTN1 A0A3P8TTR2 A0A091TAZ0 A0A091V6V0 A0A016RTJ8 A0A091RZU2 A0A016RU51
S4PS67 A0A023F8B4 A0A0V0GF60 A0A069DNF8 A0A224XMQ2 A0A0P4VNE1 R4G3X0 A7SXH9 V9LDM0 K4G087 A0A2D0RS50 A0A091R9N1 A0A1A8H3I7 A0A1A8EQS7 A0A1A8MMY7 A0A1A8NDK8 A0A1A8NEX9 A0A1A8S930 A0A3N0XE49 A0A1A8IFV7 A0A1A8JNB7 A0A1A8UKI1 A0A3Q2TVU9 W5KHX1 A0A1A8DJ81 A0A146XFF7 A0A1A7ZC24 A0A1A8CN82 A0A210QSX9 A0A3Q3A060 A0A3B5JYJ6 A0A3S1BB47 R0JWL8 A0A1V4IFV8 A0A3Q2CJI7 H3C699 A0A2R7WCX4 A0A3M6TQY6 A0A163DF35 R7VWK2 V3Z517 A0A3B3D9G8 A0A087V7E0 A0A1A7XF10 A0A087V1A5 A0A131Y815 A0A3B4C3Q8 A0A093CRB9 A0A3Q3GKT4 A0A044S4L3 A0A3P8X1E4 G1N663 A0A3Q1G8G5 A0A093NAX7 A0A087QTH5 A0A2U9BM96 A0A226NLT4 G3WT92 A0A091LHX4 A0A091IMZ1 A0A091P8D4 A0A091R091 A0A2P4SL83 A0A091SW12 A0A3B3U7C6 A0A091NKN4 G1KF12 G3MT35 A0A023GA07 A0A093BK81 W5NFL4 A0A091U9H8 A0A093KAW7 A0A226P6L2 K7FA78 A0A093II94 A0A023G795 E1BRN1 A0A293LAH9 A0A091FUX2 A0A0N4T061 A0A2I0U7D6 A0A0K0ILX7 A0A0R3R313 A0A093J7A2 A0A1S2ZL65 A0A093FY15 A0A224YTN1 A0A3P8TTR2 A0A091TAZ0 A0A091V6V0 A0A016RTJ8 A0A091RZU2 A0A016RU51
Pubmed
EMBL
NWSH01000679
PCG74843.1
KQ461155
KPJ08235.1
ODYU01003015
SOQ41250.1
+ More
KQ459232 KPJ02697.1 GDQN01005230 JAT85824.1 AGBW02010278 OWR48818.1 GAIX01014108 JAA78452.1 GBBI01001141 JAC17571.1 GECL01000189 JAP05935.1 GBGD01003286 JAC85603.1 GFTR01002649 JAW13777.1 GDKW01002140 JAI54455.1 ACPB03016231 GAHY01001850 JAA75660.1 DS469888 EDO31592.1 JW878129 AFP10646.1 JX052608 AFK10836.1 KK696155 KFQ24497.1 HAEC01009084 SBQ77300.1 HAEB01002978 SBQ49505.1 HAEF01017044 SBR58203.1 HAEG01002386 SBR67165.1 HAEH01001798 SBR67625.1 HAEI01011989 SBS14457.1 RJVU01079719 ROI15603.1 HAED01009825 SBQ96037.1 HAEE01000824 SBR20840.1 HAEJ01007639 SBS48096.1 HAEA01004826 SBQ33306.1 GCES01045435 JAR40888.1 HADY01001882 SBP40367.1 HADZ01016577 SBP80518.1 NEDP02002059 OWF51809.1 RQTK01000458 RUS79330.1 KB743061 EOB01667.1 LSYS01009753 OPJ58740.1 KK854560 PTY16880.1 RCHS01003129 RMX43823.1 KV440987 OAD70800.1 KB375613 AKCR02000106 EMC83652.1 PKK20596.1 KB203251 ESO85798.1 KL482768 KFO08532.1 HADW01015030 HADX01012710 SBP16430.1 KL820070 KFM83394.1 GEFM01000427 JAP75369.1 KL466696 KFV17018.1 CMVM020000052 KL224622 KFW62118.1 KL225899 KFM04529.1 CP026250 AWP05101.1 MCFN01000017 OXB68348.1 AEFK01032415 KL327562 KFP56818.1 KK500568 KFP08903.1 KK654807 KFQ04142.1 KK806280 KFQ32321.1 PPHD01037998 POI24875.1 KK486736 KFQ62195.1 KL387196 KFP90323.1 JO845036 AEO36653.1 GBBM01005685 JAC29733.1 KN125662 KFU83703.1 AHAT01034933 KK423389 KFQ87131.1 KL206950 KFV87459.1 AWGT02000150 OXB75242.1 AGCU01016236 KK594737 KFW02430.1 GBBM01005719 JAC29699.1 AADN05000005 GFWV01000206 MAA24936.1 KL447392 KFO72994.1 UZAD01000115 VDN82651.1 KZ506056 PKU41931.1 LN857024 CDQ01839.1 UZAG01019131 VDO42492.1 KK570061 KFW06864.1 KL214756 KFV61723.1 GFPF01007953 MAA19099.1 KK443674 KFQ71012.1 KK733733 KFQ98514.1 JARK01001714 EYB81606.1 KK938264 KFQ48626.1 KE125101 EPB71601.1 EYB81607.1
KQ459232 KPJ02697.1 GDQN01005230 JAT85824.1 AGBW02010278 OWR48818.1 GAIX01014108 JAA78452.1 GBBI01001141 JAC17571.1 GECL01000189 JAP05935.1 GBGD01003286 JAC85603.1 GFTR01002649 JAW13777.1 GDKW01002140 JAI54455.1 ACPB03016231 GAHY01001850 JAA75660.1 DS469888 EDO31592.1 JW878129 AFP10646.1 JX052608 AFK10836.1 KK696155 KFQ24497.1 HAEC01009084 SBQ77300.1 HAEB01002978 SBQ49505.1 HAEF01017044 SBR58203.1 HAEG01002386 SBR67165.1 HAEH01001798 SBR67625.1 HAEI01011989 SBS14457.1 RJVU01079719 ROI15603.1 HAED01009825 SBQ96037.1 HAEE01000824 SBR20840.1 HAEJ01007639 SBS48096.1 HAEA01004826 SBQ33306.1 GCES01045435 JAR40888.1 HADY01001882 SBP40367.1 HADZ01016577 SBP80518.1 NEDP02002059 OWF51809.1 RQTK01000458 RUS79330.1 KB743061 EOB01667.1 LSYS01009753 OPJ58740.1 KK854560 PTY16880.1 RCHS01003129 RMX43823.1 KV440987 OAD70800.1 KB375613 AKCR02000106 EMC83652.1 PKK20596.1 KB203251 ESO85798.1 KL482768 KFO08532.1 HADW01015030 HADX01012710 SBP16430.1 KL820070 KFM83394.1 GEFM01000427 JAP75369.1 KL466696 KFV17018.1 CMVM020000052 KL224622 KFW62118.1 KL225899 KFM04529.1 CP026250 AWP05101.1 MCFN01000017 OXB68348.1 AEFK01032415 KL327562 KFP56818.1 KK500568 KFP08903.1 KK654807 KFQ04142.1 KK806280 KFQ32321.1 PPHD01037998 POI24875.1 KK486736 KFQ62195.1 KL387196 KFP90323.1 JO845036 AEO36653.1 GBBM01005685 JAC29733.1 KN125662 KFU83703.1 AHAT01034933 KK423389 KFQ87131.1 KL206950 KFV87459.1 AWGT02000150 OXB75242.1 AGCU01016236 KK594737 KFW02430.1 GBBM01005719 JAC29699.1 AADN05000005 GFWV01000206 MAA24936.1 KL447392 KFO72994.1 UZAD01000115 VDN82651.1 KZ506056 PKU41931.1 LN857024 CDQ01839.1 UZAG01019131 VDO42492.1 KK570061 KFW06864.1 KL214756 KFV61723.1 GFPF01007953 MAA19099.1 KK443674 KFQ71012.1 KK733733 KFQ98514.1 JARK01001714 EYB81606.1 KK938264 KFQ48626.1 KE125101 EPB71601.1 EYB81607.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000015103
UP000001593
+ More
UP000221080 UP000265000 UP000018467 UP000242188 UP000264800 UP000005226 UP000271974 UP000190648 UP000265020 UP000007303 UP000275408 UP000077315 UP000053872 UP000030746 UP000261560 UP000054359 UP000261440 UP000261660 UP000024404 UP000265120 UP000001645 UP000257200 UP000054081 UP000053286 UP000246464 UP000198323 UP000007648 UP000053119 UP000261500 UP000001646 UP000018468 UP000053584 UP000198419 UP000007267 UP000000539 UP000053760 UP000038020 UP000278627 UP000006672 UP000050602 UP000079721 UP000053875 UP000265080 UP000053605 UP000024635
UP000221080 UP000265000 UP000018467 UP000242188 UP000264800 UP000005226 UP000271974 UP000190648 UP000265020 UP000007303 UP000275408 UP000077315 UP000053872 UP000030746 UP000261560 UP000054359 UP000261440 UP000261660 UP000024404 UP000265120 UP000001645 UP000257200 UP000054081 UP000053286 UP000246464 UP000198323 UP000007648 UP000053119 UP000261500 UP000001646 UP000018468 UP000053584 UP000198419 UP000007267 UP000000539 UP000053760 UP000038020 UP000278627 UP000006672 UP000050602 UP000079721 UP000053875 UP000265080 UP000053605 UP000024635
Pfam
PF06372 Gemin6
SUPFAM
SSF50182
SSF50182
ProteinModelPortal
A0A2A4JTP8
A0A194QTA5
A0A2H1VKA0
A0A194QB57
A0A1E1WFQ1
A0A212F502
+ More
S4PS67 A0A023F8B4 A0A0V0GF60 A0A069DNF8 A0A224XMQ2 A0A0P4VNE1 R4G3X0 A7SXH9 V9LDM0 K4G087 A0A2D0RS50 A0A091R9N1 A0A1A8H3I7 A0A1A8EQS7 A0A1A8MMY7 A0A1A8NDK8 A0A1A8NEX9 A0A1A8S930 A0A3N0XE49 A0A1A8IFV7 A0A1A8JNB7 A0A1A8UKI1 A0A3Q2TVU9 W5KHX1 A0A1A8DJ81 A0A146XFF7 A0A1A7ZC24 A0A1A8CN82 A0A210QSX9 A0A3Q3A060 A0A3B5JYJ6 A0A3S1BB47 R0JWL8 A0A1V4IFV8 A0A3Q2CJI7 H3C699 A0A2R7WCX4 A0A3M6TQY6 A0A163DF35 R7VWK2 V3Z517 A0A3B3D9G8 A0A087V7E0 A0A1A7XF10 A0A087V1A5 A0A131Y815 A0A3B4C3Q8 A0A093CRB9 A0A3Q3GKT4 A0A044S4L3 A0A3P8X1E4 G1N663 A0A3Q1G8G5 A0A093NAX7 A0A087QTH5 A0A2U9BM96 A0A226NLT4 G3WT92 A0A091LHX4 A0A091IMZ1 A0A091P8D4 A0A091R091 A0A2P4SL83 A0A091SW12 A0A3B3U7C6 A0A091NKN4 G1KF12 G3MT35 A0A023GA07 A0A093BK81 W5NFL4 A0A091U9H8 A0A093KAW7 A0A226P6L2 K7FA78 A0A093II94 A0A023G795 E1BRN1 A0A293LAH9 A0A091FUX2 A0A0N4T061 A0A2I0U7D6 A0A0K0ILX7 A0A0R3R313 A0A093J7A2 A0A1S2ZL65 A0A093FY15 A0A224YTN1 A0A3P8TTR2 A0A091TAZ0 A0A091V6V0 A0A016RTJ8 A0A091RZU2 A0A016RU51
S4PS67 A0A023F8B4 A0A0V0GF60 A0A069DNF8 A0A224XMQ2 A0A0P4VNE1 R4G3X0 A7SXH9 V9LDM0 K4G087 A0A2D0RS50 A0A091R9N1 A0A1A8H3I7 A0A1A8EQS7 A0A1A8MMY7 A0A1A8NDK8 A0A1A8NEX9 A0A1A8S930 A0A3N0XE49 A0A1A8IFV7 A0A1A8JNB7 A0A1A8UKI1 A0A3Q2TVU9 W5KHX1 A0A1A8DJ81 A0A146XFF7 A0A1A7ZC24 A0A1A8CN82 A0A210QSX9 A0A3Q3A060 A0A3B5JYJ6 A0A3S1BB47 R0JWL8 A0A1V4IFV8 A0A3Q2CJI7 H3C699 A0A2R7WCX4 A0A3M6TQY6 A0A163DF35 R7VWK2 V3Z517 A0A3B3D9G8 A0A087V7E0 A0A1A7XF10 A0A087V1A5 A0A131Y815 A0A3B4C3Q8 A0A093CRB9 A0A3Q3GKT4 A0A044S4L3 A0A3P8X1E4 G1N663 A0A3Q1G8G5 A0A093NAX7 A0A087QTH5 A0A2U9BM96 A0A226NLT4 G3WT92 A0A091LHX4 A0A091IMZ1 A0A091P8D4 A0A091R091 A0A2P4SL83 A0A091SW12 A0A3B3U7C6 A0A091NKN4 G1KF12 G3MT35 A0A023GA07 A0A093BK81 W5NFL4 A0A091U9H8 A0A093KAW7 A0A226P6L2 K7FA78 A0A093II94 A0A023G795 E1BRN1 A0A293LAH9 A0A091FUX2 A0A0N4T061 A0A2I0U7D6 A0A0K0ILX7 A0A0R3R313 A0A093J7A2 A0A1S2ZL65 A0A093FY15 A0A224YTN1 A0A3P8TTR2 A0A091TAZ0 A0A091V6V0 A0A016RTJ8 A0A091RZU2 A0A016RU51
Ontologies
GO
PANTHER
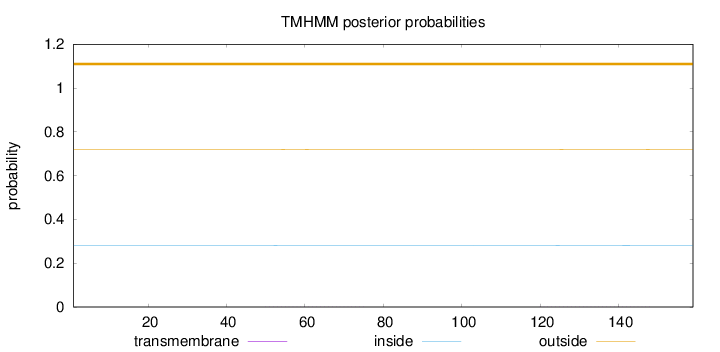
Topology
Length:
159
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03731
Exp number, first 60 AAs:
0.00666
Total prob of N-in:
0.28085
outside
1 - 159
Population Genetic Test Statistics
Pi
188.971803
Theta
221.508697
Tajima's D
-0.444021
CLR
0
CSRT
0.256937153142343
Interpretation
Uncertain
